Exporting GIF of Time-Series Trend Analysis from Kriging
Contents
Introduction
Purpose
Animating data has the ability to engage viewers in ways numerical data cannot, and can demonstrate how a phenomenon is connected and evolves through a series of events. The ability to visualize meteorological data via a time series animation, provides viewers with an option to see the progression of cold fronts, heat waves and precipitation, and can assist in forecasting events allowing agriculture specialist to plan crop planting, irrigation, harvesting and chemical application. In the example that will be demonstrated in this tutorial, precipitation data will be used to create a time series event that would be useful in reviewing historical data that can be used in cross analyzing weather events with floods or droughts, and to detect patterns.
Jupyter Lab will be the platform used to create the time series animation, in conjunction with Anaconda, Pandas, and PyKrige. Using Jupyter Lab will enable the user to develop skills in scripting, and is a more efficient way to complete our time series animation. Jupyter is a flexible open-source environment for notebooks, code and data that is capable of handling a wide variety of file formats. Anaconda will provide the building environment to enhance ease of collaborative work and to ensure a homogeneous work environment. Anaconda is preferred for our tutorial due its ease of use for beginner programmers with its pre-installed packages that that are helpful for data science and machine learning. Pandas is a package built on top of another package (Numpy), and will facilitates data analysis by providing python functions and methods (specifically for Time Series creation); and lastly PyKrige provides a Python library that is used for geostatistical tasks such as interpolation, which we will use to create new points to provide a smoother time series animation.
The intended audience of this tutorial are those that existing knowledge and experience using Python or Anaconda, Notebooks (CoLab or Jupyter) and Pandas.
Environment
For this tutorial, we will be working from Windows 10. Most steps, except the installation of software, can be used in a Linux GUI.
Procedure
We will take the following steps in this tutorial to accomplish our goal. When we are done, we will have an animated GIF showing the monthly precipitation in Ontario from 2000 to 2010, inclusively.
Acquiring Data
The data we will use in this tutorial is the Adjusted and Homogenized Canadian Climate Data (AHCCD). It can be downloaded here: https://climatedata.ca/download/
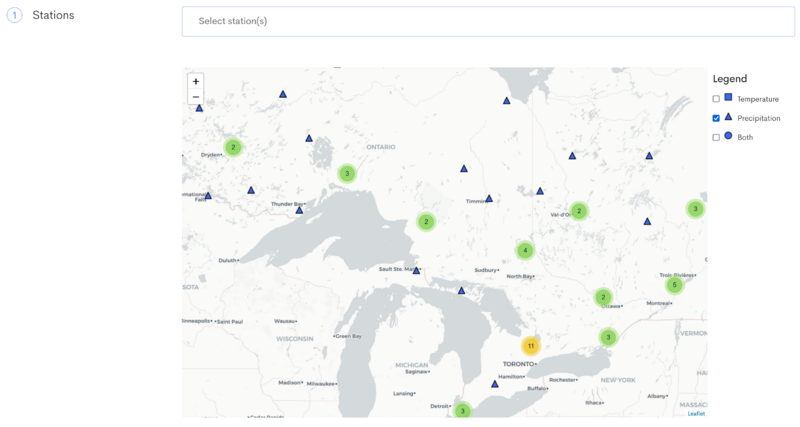
Click on the AHCCD tab, and scroll down to the map where you can select the stations you want to download data from.
On the right side, deselect 'Temperature' and 'Both', and leave only "Precipitation" selected.
Next, select all stations in Ontario by left-clicking each one.
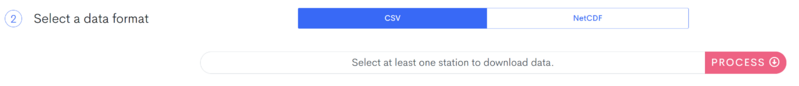
Finally, scroll down and set the data type to CSV and click the red 'Process' button.
Download and Install Anaconda
Download and install Anaconda for Windows.
Anaconda can be downloaded here: https://www.anaconda.com/
The installation instructions can be found here: https://docs.anaconda.com/anaconda/install/windows/
Setup an Anaconda Environment
From the Start Menu, run "Anaconda Command Prompt"
Type the following command to create your Anaconda Environment.
conda create -n gis python jupyter
This line will create a conda environment with the name gis, and python and jupyter already installed.
Now let's activate that environment. You will notice that before activating it, the left side of the command line shows "(base)" indicating that you are in the "base" environment.
conda activate gisNow it should read "(gis)" to indicate that you are in the "gis" environment.
Next, we'll install some required modules for our workflow. Type in the following commands.
pip install rasterio pykrige pandas geopandas matplotlib numpy jupyterlabFinally, we need to download and install GDAL. We will use Christoph Gohlke's prepared wheel file. Follow this link and download the latest version of GDAL. https://www.lfd.uci.edu/~gohlke/pythonlibs/
After downloading, navigate to the folder containing the downloaded file from the Anaconda Command Prompt.
For example, if the file is in C:\Users\GIS_User\Downloads, you will enter the command as follows:
cd C:\Users\GIS_User\DownloadsThen, run the following command to install GDAL using pip. You can type the first few letters of the filename you downloaded, and press TAB to autocomplete the rest.
pip install GDAL-3.4.3-cp310-cp310-win_amd64.whlNote that your filename may be different. You will need to use the exact filename of the file you downloaded.
Run Jupyter Lab and Create a Jupyter Notebook
Create a new working folder and place the AHCCD.CSV file into it. From the same Anaconda Command Prompt, navigate to the folder you created. We can launch the Jupyter Notebook by typing the following command.
jupyter labJupyter Lab will launch into your browser. It may prompt you which browser you would like it to launch to the first time you run this command.
After working in Jupyter Lab and saving your work, you may want to return to it another day and continue from where you left off. You can do this by activating the conda environment and using the same command to launch jupyter lab.
To do this, launch the Anaconda Command Prompt and type in the following commands. Don't forget to navigate to your working folder first.
conda activate gis
jupyter lab
Create a new notebook and name it as you would like. In the Jupyter notebook, you have blocks that can be set to Markdown or Code. The Code blocks will use Python code.
Create a code block and enter the following code in it.
from osgeo import gdal
import os
import pandas as pd
import imageio.v2 as imageio
from matplotlib import pyplot as plt
import matplotlib as mpl
from pykrige.ok import OrdinaryKriging
import numpy as np
import warnings
warnings.filterwarnings("ignore", category=FutureWarning)
This code will import the required modules, and set the output to ignore warning pertaining to future changes in the modules.
Create another code block below this one and enter the following code.
# Open CSV dataframe with pandas.
df = pd.read_csv('ahccd.csv')
This code block will open our csv file as a pandas dataframe. We can now begin to manipulate the dataframe and pull the relevant data from it.
Create another code block and enter this code. It will show us the data types within our dataframe.
# Get the data types from the dataframe.
df.dtypes
Here is the code for the next code block. This will show us the 'time' column of the first several rows and the last several rows of our dataframe. We can see the date range we expect to see in our data.
# Get the date range from the 'time' column.
print(df['time'])
We want to be able to filter by year, and by month. To do this easily, we will take the yyyy-mm-dd format of the 'time' column and split it into three new columns named year, month, and day.
# Replace all empty field with a zero.
df['pr'] = df['pr'].fillna(0)
# Create three new columns from the 'time' column to store the year, month, and day separately.
df[['year','month','day']] = df['time'].str.split('-',expand=True) #Split the string on "-" and put the results in 3 new columns
We will need to convert these new columns to int (integer type).
# Convert the three new columns to int.
df['year'] = df['year'].astype('int')
df['month'] = df['month'].astype('int')
df['day'] = df['day'].astype('int')
Now we can select a subset of the data from the year 2000 to 2010, inclusively, and select only the data in the months of April through September.
# Select a subset of the dataframe, where the year is between 2000 and 2010 inclusively, and the month is between 4 and 9 inclusively.
sel = df[(df['year']>=2000) & (df['year']<=2010) & (df['month']>=4) & (df['month']<=9)]
We now have a smaller and more manageable dataframe. Let's export it to a new CSV file for future use.
# Export the selection to a new CSV file.
sel.to_csv('precip_2000-2010_ON.csv')
Let's also check that the data is what we expect. We can get the unique year values from the data to verify.
# Print the unique year values to verify if the correct subset was selected.
print(sel['year'].unique())
The output from that last code block should have yielded a list of years from 2000 to 2010.
After verifying the data is what we expect, we can take a look at the column names to decide which are relevant for our workflow.
# Print the column names.
print(sel.columns)
To shorten processing time, we can exclude the columns that we don't need. This next code block will go over several steps.
The comments in the code will explain the steps taken. The end result is two numpy arrays of longitudes and latitudes representing the coordinates of each station, and a numpy array of precipitation values.
These will be required for the kriging.
# Make a list of the desired columns.
desired_columns = ['time', 'year', 'month', 'station_name', 'pr', 'elev', 'lon', 'lat']
# Make lists from the unique year, month, and station values.
years = sel['year'].unique()
months = sel['month'].unique()
stations = sel['station_name'].unique()
# Create a new dataframe containing the desired columns. We will input data from our selected subset.
means = []
# Create lists for longitudes, latitudes, and precipitation data. These will be populated in the for loops below.
lons = []
lats = []
data = []
# Keep track of min and max mean precipitation values
min_pr = 0
max_pr = 0
# We have daily values, and we would like monthly values instead. Select a subset of data for each month, at each station, and calculate its mean value.
# Then, store the mean values into the means dataframe.
for year in years:
for month in months:
station_monthly_means = pd.DataFrame(columns = desired_columns)
for station in stations:
sbst = sel[desired_columns]
sbst2 = sbst[(sbst['year'] == year) & (sbst['month'] == month) & (sbst['station_name'] == station)]
# get mean for each station and add to means df
station_monthly_means = station_monthly_means.append(sbst2.iloc[0]).reset_index(level=0, drop=True)
station_monthly_means.loc[station_monthly_means.index[-1], 'pr'] = sbst2['pr'].mean()
if min_pr == 0:
min_pr = sbst2['pr'].mean()
if sbst2['pr'].mean() < min_pr:
min_pr = sbst2['pr'].mean()
if sbst2['pr'].mean() > max_pr:
max_pr = sbst2['pr'].mean()
means.append(station_monthly_means)
# Append the longitudes, latitudes, and data from all stations per month
lons.append(np.array(means[-1]['lon']))
lats.append(np.array(means[-1]['lat']))
data.append(np.array(means[-1]['pr']))
# Print the first row from each of the first 5 months to verify.
for month in means[:5]:
print(month.head(1))
Next we prepare grid data that will be required for Kriging.
# The size of each square in the grid.
grid_space = 0.1
# Since we're putting together a timeseries, we'll make a list of grids for our longitudes, latitudes.
grid_lons = []
grid_lats = []
# Create lists for our grid. This is required for the Kriging steps.
for lon in lons:
grid_lons.append(np.arange(-100, -70, grid_space, dtype='float64')) #grid_space is the desired delta/step of the output array
for lat in lats:
grid_lats.append(np.arange(37, 60, grid_space, dtype='float64'))
The following step is where we perform the Kriging. Since we're working with a time series, we will run Kriging once for every month, where the precipitation values from each station for that month are used to create the interpolation result.
# We'll store our results in lists as well.
z_list = []
ss_list = []
# We will need to use an index to access lons and lats that correspond with each data point.
# NOTE: This will take a LONG time! Go stretch for a while.
for i in range(len(data)):
OK = OrdinaryKriging(lons[i], lats[i], data[i], variogram_model='gaussian', verbose=True, enable_plotting=False,nlags=150, weight=True)
z1, ss1 = OK.execute('grid', grid_lons[i], grid_lats[i], backend='loop')
z_list.append(z1)
ss_list.append(ss1)
Note that the output is quite long. This tutorial is focused on how to use the tools rather than on how to create the best results using these tools. You will noticed that the nugget results vary greatly, and this will affect the final GIF.
As this tutorial is only designed to show how these tools can be used, it is up to you to read through the PyKrige documentation and find the best way to get accurate results from your data. You can read the documentation here: https://geostat-framework.readthedocs.io/projects/pykrige/en/stable/
Specifically, you can explore the different functions. We use Ordinary Kriging in this tutorial, but you can try other functions from the PyKrige module. You will find that the documentation shows the parameters of each function. You can see the expected inputs, whether they are mandatory or optional, and the expected results. Optional inputs will have a default value (i.e. input=value). You can also see the expected types for both inputs and returns (outputs).
We will now create a custom colormap to use in our figures. The custom colormap takes in a dictionary of colours. These are always red, green, and blue. The matplotlib documentation will show that this dictionary has the following format:
cdict = { 'red' : [(0.0, 0.0, 0.0),<br>
(0.5, 1.0, 1.0),<br>
(1.0, 1.0, 1.0)],<br>
'green': [(0.0, 0.0, 0.0),<br>
(0.5, 0.0, 0.0),<br>
(0.75, 1.0, 1.0),<br>
(1.0, 1.0, 1.0)],<br>
'blue' : [(0.0, 0.0, 0.0),<br>
(0.5, 0.0, 0.0),<br>
(1.0, 1.0, 1.0)]}<br>The documentation states that this should show you a gradient made up of red in the bottom half, green in the middle half, and blue in the top half. The best way to interpret this information is by picturing a rectangle, and adding red to the bottom 50% of the rectangle, then adding green to the area that covers from 25% up from the bottom of the rectangle to 75%, and finally adding blue to the top half of the rectangle. There will be some overlap where half the red portion is mixed with green, and half the green portion is mixed with blue. To set a custom colour, we will look at the list of values for each colour.
'red' is shown with three sets of numbers, and each set has three numbers separated by commas. These are scalar values ranging from 0 to 1. These sets represent the location, the value, and the alpha value. The first set reads (0.0, 0.0, 0.0), meaning (0.0 = bottom, 0.0 value = no red, 0.0 alpha = transparent). The next set is (0.5, 1.0, 1.0) meaning (0.5 = halfway, 1.0 value = full red, 1.0 alpha = opaque). The values from the first set trend towards the values from the second set in a linear fashion creating a gradient, and so on in that fashion with subsequent sets. This means we can mix RGB colors at different locations to create custom colors, and use the placements to create custom gradients. Here is the code for a custom colormap suitable for visualizing precipitation values.
# Create a custom color gradient for our colormap.
cdict = { 'red' : [(0.0, 1.0, 1.0),
(0.05, 0.59, 0.59),
(1.0, 0.0, 0.0)],
'green': [(0.0, 1.0, 1.0),
(0.05, 0.29, 0.29),
(0.2, 1.0, 1.0),
(1.0, 0.0, 0.0)],
'blue' : [(0.0, 0.88, 0.88),
(0.05, 0.1, 0.1),
(0.2, 1.0, 1.0),
(1.0, 1.0, 1.0)]}
cm = mpl.colors.LinearSegmentedColormap('my_colormap', cdict, 1024)
With our custom colormap created, we will create our figures and export them to PNGs.
# Create a list of month labels
months=['Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep']
# For each set of precipiation values, we will clip the Kriging values to the Ontario boundaries
# and create a figure and export it to a png file.
for i in range(len(z_list)):
ras = gdal.Translate('temp.tif', gdal_array.OpenArray(z_list[i]), format = 'MEM',
outputSRS = 'EPSG:4326', outputBounds = [-100, 60, -70, 37])
ras2 = gdal.Warp('temp2.tif', ras, format='MEM',
options=' -cutline lpr_000b16a_e.shp' + ' -dstalpha')
cax = plt.imshow(np.flipud(ras2.GetRasterBand(1).ReadAsArray()), cmap=cm, vmin=min_pr, vmax=max_pr, extent=(-100, -70, 37, 60), origin='lower')
plt.scatter(lons[i], lats[i], s=0.1, c='k', marker='.')
cbar=plt.colorbar(cax, label='Precip. (mm)')
# Label the figure with the year and month for easy identification.
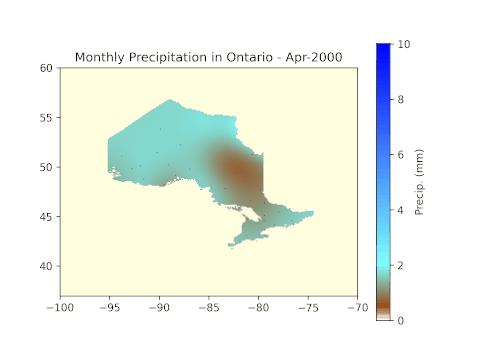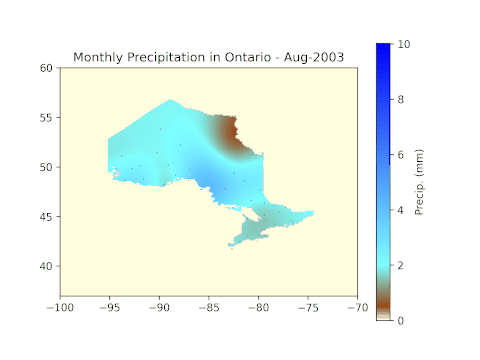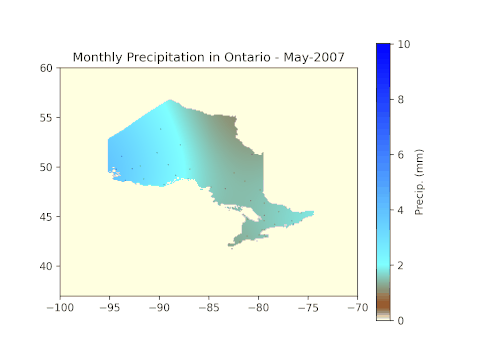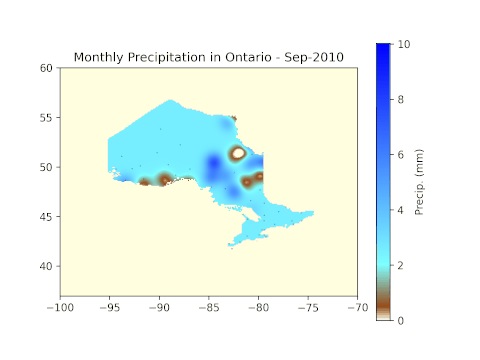
plt.title('Monthly Precipitation in Ontario - ' + months[means[i]['month'][0] - 4] + '-' + str(means[i]['year'][0]))
# Export figures to png files.
if (not os.path.isdir('pngs')):
os.mkdir('pngs')
plt.savefig('pngs/'+ str(i) + '.png',dpi=300)
plt.close()
Finally, we'll take the PNGs and create a GIF out of them.
# build gif
with imageio.get_writer('precip.gif', mode='I', duration = 0.4) as writer:
for i in range(len(os.listdir('./pngs/'))):
if filename[-4:] != '.png':
continue
image = imageio.imread('pngs/' + str(i) + '.png')
writer.append_data(image)
Results
After running all code blocks in the Jupyter Notebook, we can expect to see the following GIF in the working folder.