Landscape Change Assessment using Unsupervised Image Classification in GRASS GIS
Contents
Purpose
The purpose of this tutorial is to assess changes to landscape composition (land-use type) and configuration (arrangement of land-use types) over a period of time. In order to assess these changes, a technique called unsupervised image classification will be used. This tutorial has been created for a user who has some experience with geographic information systems (GIS) software and is somewhat familiar with using raster data. In theory, the user should have some background (or at least some interest) in landscape ecology and will be able to apply their findings to problems with some spatio-temporal aspects.
Introduction
Unsupervised classification is a method that categorizes pixels (or cells) into classes based on a certain characteristic using an algorithm. Alternatively, supervised classification requires the user to specify the classes based on some characteristic, and uses this information as an input which trains software to classify pixels. In this tutorial, the characteristic of interest is the spectral reflectance of different land-use types.
To capture an accurate image of what land-use types exist, remote sensing data is used because it captures images and information across a spectrum of wavelengths called bands. Each of these bands contain different information about what is on the ground because different land-use types absorb and reflect wavelengths at different. This tutorial specifically uses Landsat 7 and Landsat 8 data which captures images across eight bands. More information about the Landsat program can be found here. In this tutorial, the “false colour” composite is used, which contains information from spectral bands 4, 3, and 2 which represent the near-infrared, red, and green bands, respectively. This combination of spectral bands allows the user to identify different classes of vegetation compared to water and urban land-use.
GRASS (Geographic Resources Analysis Support System) GIS is open-source software that is free to download and use. The software is commonly used for image processing, spatial analysis and modelling, and visualization of geospatial data. It can be used on its own, or in combination with other software such as QGIS and R. GRASS GIS is a founding member of the Open Source Geospatial (OSGeo) Foundation along with other open source projects. More information about GRASS GIS can be found [ https://grass.osgeo.org/ here] and more information about OSGeo can be found here.
Software Download and Installation
In this tutorial, GRASS GIS version 7.8.1 for Windows is being used, however this software is also available for Mac and Linux users. The latest version of GRASS GIS may be downloaded here. This website contains very useful information and documentation for users that have never been exposed to GRASS GIS before now. I highly suggest looking at this documentation, as well as this tutorial which provides more information on GRASS GIS. Throughout this tutorial, I will reference different sections of the GRASS GIS Manual which contains lots of helpful information.
Data Set
Because this tutorial is being written to demonstrate the ability of GRASS GIS to assess changes in landscapes, a data set has been provided for the user. There are many sources where one can find satellite imagery data, however a data set is being provided to the user so they may follow along with this tutorial without having their own data set. This data set consists of four GeoTiff files acquired using the Earth Observing System (EOS) Landviewer tool. Two of these GeoTiff files contains an image obtained by Landsat 7 on September 7, 2000 and an image obtained by Landsat 8 on September 23, 2017 using the false-colour composite, while the other two images use the visible light bands (1, 2, 3). Both images contain imagery of the same area of interest (AOI) which consists of a 40.04 km² square area centered at 45.03001°N 75.68135°W with 30m resolution. This area of interest is located in Kemptville, Ontario, an area which has undergone extensive development since the early 2000s to accommodate increased population. The population of Kemptville is expected to continue to rise given its close proximity to Ottawa, making it an ideal location for commuters to reside, which means that further development is also expected.
To download the imagery required for this tutorial, click on the links in the table below. This will begin downloading each of the files to your computer.
| Landscape Change Assessment Data Set | |
|---|---|
| Data Information | Data File |
| GeoTIFF file for imagery from September 7, 2000 with bands 2, 3, and 4 (Landsat 7) | September 2000 File |
| GeoTIFF file for imagery from September 23, 2017 with bands 2, 3, and 4 (Landsat 8) | September 2017 File |
| GeoTIFF file for imagery from September 7, 2000 with bands 1, 2, and 3 (Landsat 7) | September 2000 Visible Bands File |
| GeoTIFF file for imagery from September 23, 2017 with bands 1, 2, and 3 (Landsat 8) | September 2017 Visible Bands File |
Methods
Starting GRASS GIS
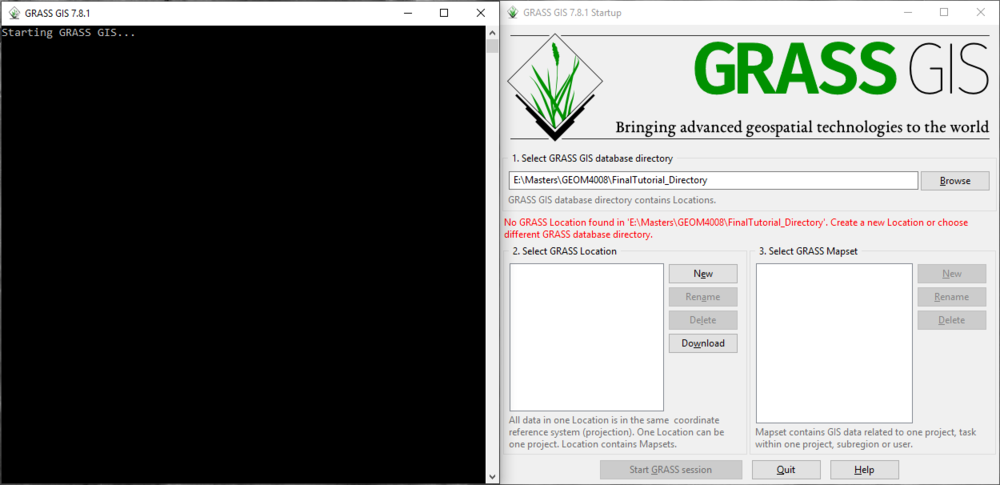
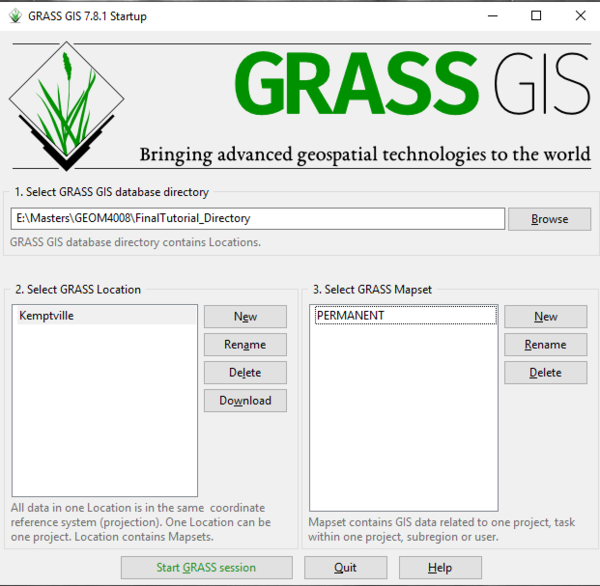
Once GRASS GIS is properly installed on your computer, you may locate it and open the program. The GRASS GIS startup interface is where the user will set their database directory and define characteristics of the project location (Figure 1). To begin using GRASS, we must first set a working (database) directory, with a project location and a mapset. The data directory is a folder on your computer where you will store all of the data for your project. The project location is a subdirectory within the database directory that contain spatial information about the project you are working on. This spatial information includes things like the coordinate system and the projection information. Within a project location, you may have multiple mapsets which contain different layers and maps that are all related to one another based on the characteristics of the project location. In this tutorial, we will use the “PERMANENT” mapset which is created automatically within the project location. More information about starting with GRASS for the first time can be found in the Quickstart Guide here.
Figure 1: The GRASS GIS Startup Interface
- 1) Click Browse under “Select GRASS GIS database directory” to navigate to the folder you wish you use as the database directory on your computer. The message seen in red font in Figure 1 should appear.
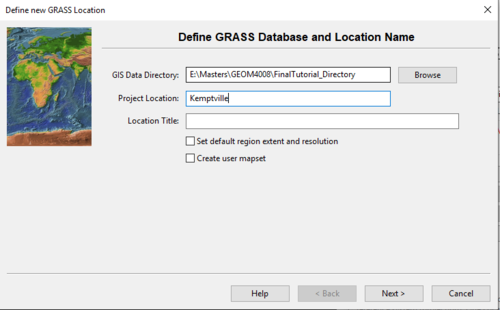
- 2) Click New under “Select GRASS Location” to create a new project location. The window seen in Figure 2 should appear.
Figure 2: The "Define New GRASS Location" window where the user selects a Data Directory and Project Location.
- 3) Choose a name for your project location and type it into the appropriate box. In this example, the name of the project location is “Kemptville”. It is best to keep the project location name simple, and if desired, a location title can be input which describes the project location in more detail.
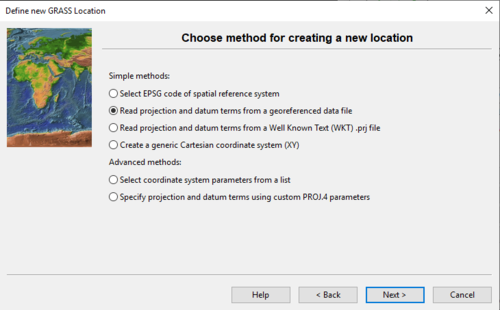
- 4) Click Next and Figure 3 should appear. In this example, we will use the second method (“Read projection and datum terms from a georeferenced data file”) to create our new project location.
Figure 3: The "Define New GRASS Location" window where the user chooses a method for creating a new Location.
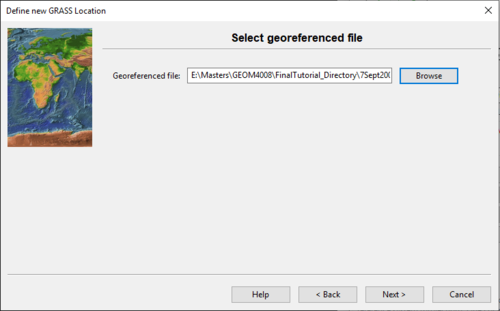
- 5) Click Next to select a georeferenced file (Figure 4).
Figure 4: The "Define New GRASS Location" window where the user selects a georeferenced file.
- 6) Click Browse and navigate to the GeoTIFF file you downloaded earlier called “7Sept2000.tiff”.
- 7) Click Next to review your selection and the projection definition on the Summary window (Figure 5.)
Figure 5: The "Define New GRASS Location" window summary page.
- 8) Click Finish to create the project location. The Startup window should now appear as it does in Figure 6.
***You may receive a pop-up at this stage that asks you if you want to import the 7Sept2000.tiff file into your newly created location. Click Yes.***
Figure 6: The "GRASS Startup" window with a Directory, Location, and Mapset selected.
- 9) Click Start GRASS session in green font at the bottom, left of the startup interface. Three windows should appear:
- The “GRASS Command” window (Figure 7). You may use this window to call on different modules (tools) or you can find them using the graphical user interface (GUI).
- The “GRASS Map Display” window (Figure 8). This is where our different map layers will appear when they are selected.
- The “GRASS Layer Manager” window (Figure 9). This is the main GUI where we will be working in this tutorial. Take notice of the different tabs along the bottom of the window (Layers, Console, Modules, Data, Python). The “Layers” tab is where the different map layers will appear when they are selected or created. The next important tab is the “Modules” tab where you can find all the modules (tools) we will use in this tutorial. The last tab we will use in this tutorial is the “Data” tab, which shows you all of the project locations and mapsets that are stored in the current database directory.
Figure 7: The "GRASS Command" window.
Figure 8: The "GRASS Map Display" window.
Figure 9: The "GRASS Layer Manager" window.
Adding Extensions
Because GRASS is open-source software, users are free to create their own extensions. These extensions are also called “add-ons”, and are modules created by users to carry out specific tasks that are not offered by any existing modules. More information about extensions, including manual pages for all of the extensions and information about developing an add-on can be found here.
There are two extensions that we will download in this tutorial. This first extension is the r.smooth.seg extension which will be used to smooth and segment our images based on spectral data, and the second is r.change.info which is used to compare classified images to one another to determine locations of change.
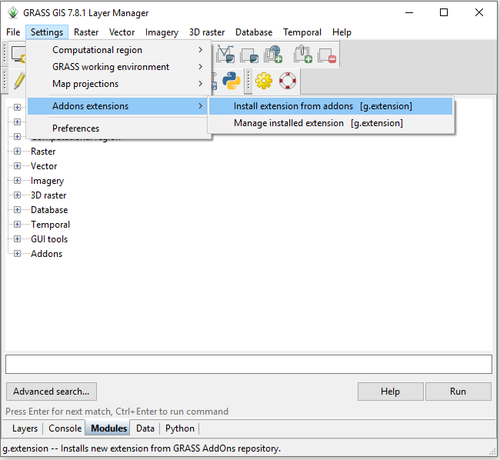
- 1) Navigate to the g.extension module by clicking on Settings > Addons Extensions > Install Extension from Addons (Figure 10).
Figure 10: The path to install new add-ons in GRASS GIS.
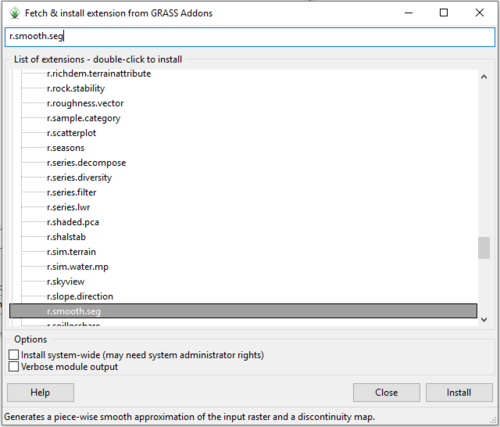
- 2) Type r.smooth.seg into the search bar at the top of the g.extension window (Figure 11).
Figure 11: The "Fetch & Install Extension from GRASS Add-ons" window with search results for the r.smooth.seg add-on.
- 3) Click on the r.smooth.seg module in the list of extensions. Notice that there is a description of what the module does in the bottom of the window.
- 4) Double-click on the extension or select it and click Install in the bottom right corner of the window to install the extension.
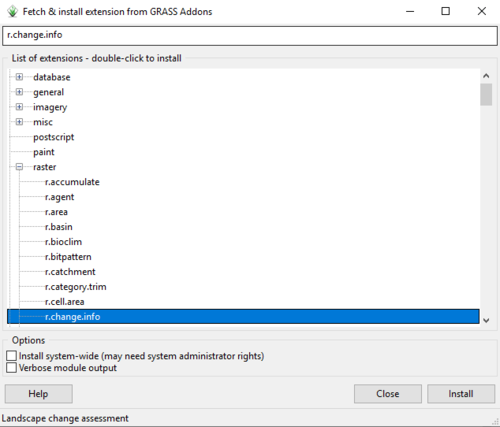
- 5) Now we will install the second extension we need by typing "r.change.info" into the search bar at the top of the g.extension window (Figure 12).
Figure 12: The "Fetch & Install Extension from GRASS Add-ons" window with search results for the r.change.info add-on.
- 6) Double-click on the extension to install it.
- 7) At this point, GRASS GIS must be restarted to load the new extensions.
- 8) Upon opening GRASS GIS for the second time, navigate to the “Modules” tab at the bottom of the "Layer Manager" window.
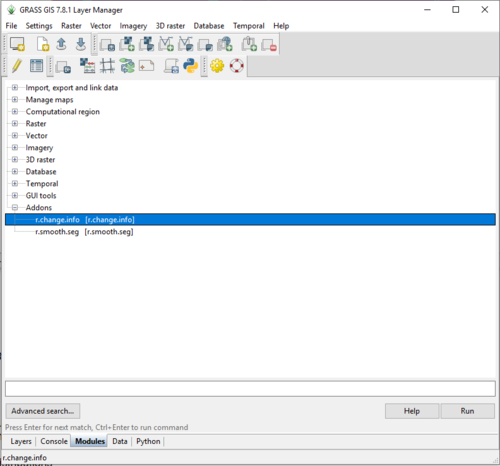
- 9) Click the + sign beside the “Addons” category to expand it (Figure 13).
- 10) To run modules through the “Modules” tab, double-click on them, or select it and click Run in the bottom right corner.
Figure 13: The "Layer Manager" window with the Modules tab open, and the newly installed add-on selected.
Importing Rasters
Now that all of the modules we need are loaded into GRASS, we can import our data. To do this, we will use the r.in.gdal module. This module imports raster data into GRASS using the GDAL library, which supports many different file formats. More information about this module can be found here. More information about the GDAL library can be found here.
Because we used the 7Sept2000.tiff file as the georeferenced file for our projection location, this file should already be imported. This can be confirmed on the “Data” tab on the “Layer Manager” window.
For the remainder of this tutorial, I will be using the 7Sept2000.tiff raster as an example in each of the steps. However, the steps in the remainder of the tutorial (apart from the Landscape Change Assessment section) will need to be carried out twice: once for our September 2000 data, and once for our September 2017 data.
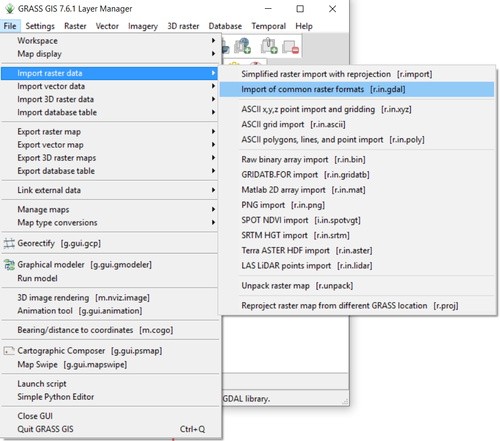
- 1) Navigate to the r.in.gdal module by clicking on File > Import Raster Data > Import of Common Raster Formats (Figure 14).
Figure 14: The path to import raster data using the r.in.gdal module.
- 2) The r.in.gdal module window will open. Notice the tabs along the top of the window, which each have different options that you can apply to your raster. The “Command Output” tab is very useful because it shows you the progress of the module and provides you with information about the module outputs. This tab shows up in all of the module windows and will be referred to in later steps.
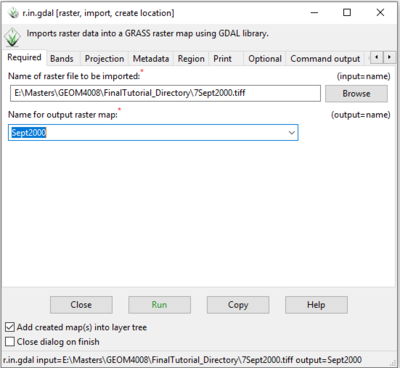
- 3) Fill in the input and output information on the “Required” tab. The input here is the location of the imagery you downloaded at the beginning of the tutorial (in this case the 7Sept2000.tiff image). The output is the name you would like the raster to have (Figure 15).
- 4) Once you have this information filled out, click the green Run button to run the module.
Figure 15: The "r.in.gdal Module" window showing input and output raster information on the "Required" tab.
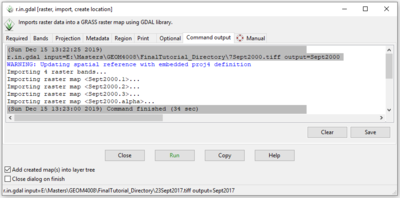
- 5) Click on the “Command Output” during/after the run to watch the progress and see information about the module output (Figure 16).
Figure 16: The "r.in.gdal Module" window showing the "Command Output" tab.
- 6) After the module is complete, repeat these steps for the 23Sept2017.tiff imagery.
- 7) Close the r.in.gdal module window and navigate to the “Data” tab on the “Layer Manager” window.
- 8) Confirm that the imagery you have just imported appears here. If the layers are not present, press the green “Refresh” button and they will appear (Figure 17). Notice that there are 4 layers imported for each image you imported. This is because GRASS separates the 3 spectral bands (2, 3, 4) imported from the image and also includes an alpha channel (Sept2000.alpha) that contains NULL cells.
Figure 17: The "Refresh" button on the "Data" tab in the "Layer Manager" window.
Visualizing Layers
In this section, we will find different ways to view the different layers in our mapset. Data visualization is extremely important for presenting data in ways that help the viewer understand what they are looking at, which allows for better interpretation of results. First, we will use the d.rgb module to view the full images in the “Map Display” window, and then we will use the g.gui.mapswipe (MapSwipe) module to view two layers at the same time. More information about the d.rgb module can be found here. More information about the the g.gui.mapswipe (MapSwipe) module can be found here.
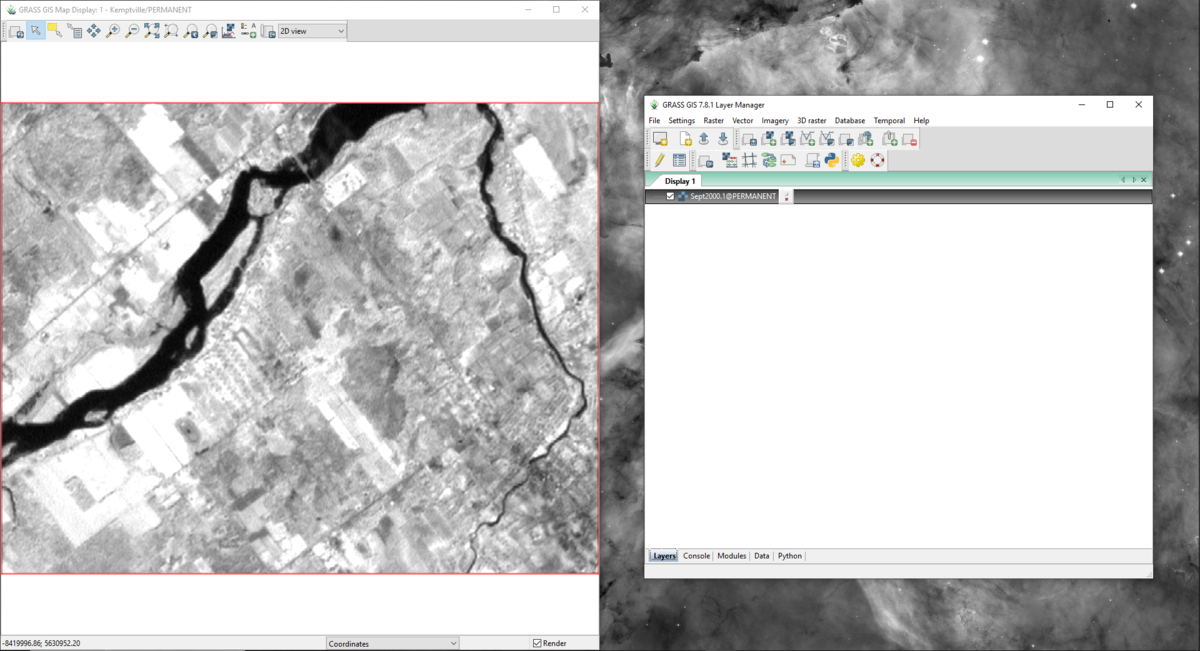
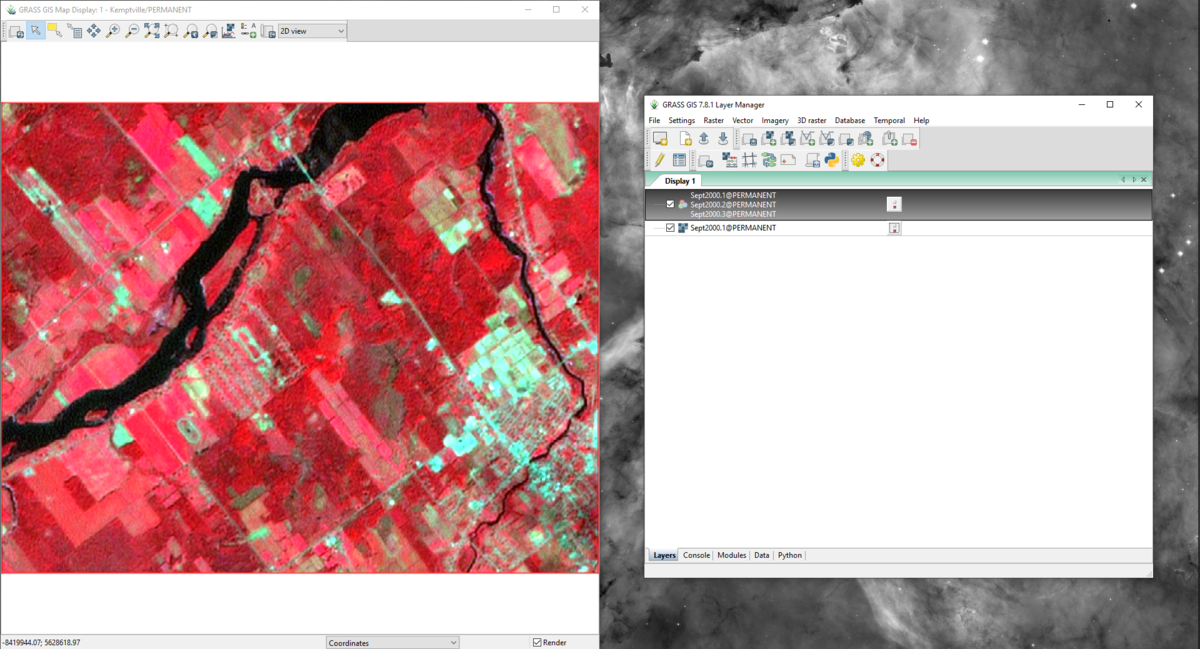
- 1) To view a layer in the “Map Display” window, double-click on the layer of interest on the “Data” tab on the “Layer Manager” window. The layer will appear on the “Layers” tab on the “Layer Manager” window and the image will appear on the “Map Display” window (Figure 18).
Figure 18: The "Map Display" window and "Layer Manager" window showing the "Sept2000.1" layer.
- 2) Navigate to the d.rgb module by following the path in Figure 19.
Figure 19: The path to add an RGB map layer using the d.rgb module.
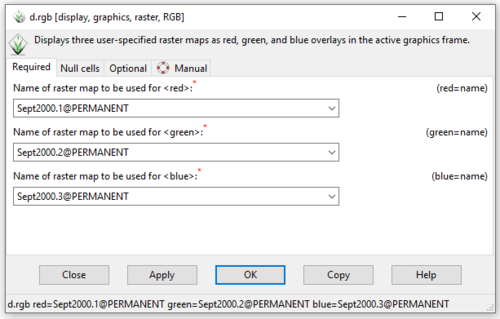
- 3) The d.rgb module will open. Because the data being used in this part of the tutorial contains spectral information from bands 2, 3, and 4 (instead of 1, 2, and 3) we will enter the bands in 1, 2, 3 order based on the name they received after importing them. This is because the near-infrared band (4) is identified as the red band (1) by the d.rgb module, and the red band (3) and green band (2) are being identified as the green (2) and blue (3) bands, respectively. This shift is reflected in our input of each of the layers in this module in Figure 20.
Figure 20: The "d.rbg Module" window showing the required input raster information.
- 4) Click Apply, then OK. You may now close this module window. The RGB layer will appear in the “Layer Manager” window and in the “Map Display” window as it does in Figure 21. If your image looks different than the one in Figure 21, it is possible that the spectral bands may have been imported in a different order. In this case, just re-arrange the order of the bands in the d.rgb module window and re-apply the change until your bands are ordered correctly and the output image looks like the one in Figure 21.
Figure 21: The RGB map layer output in the "Map Display" window and the selected RGB map layer in the "Layer Manager" window.
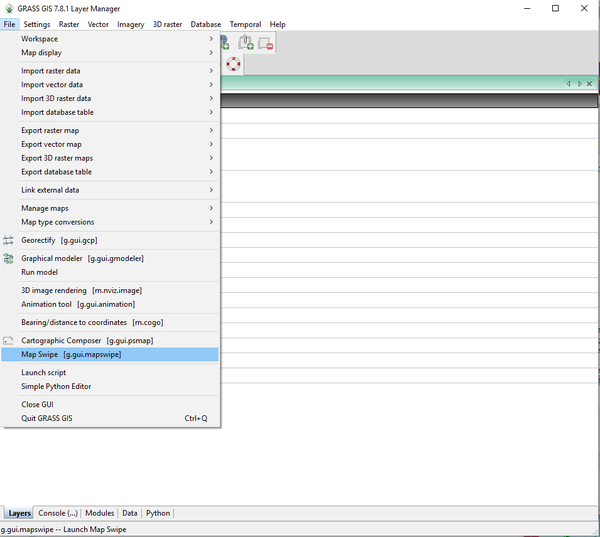
- 5) Now we will use the g.gui.mapswipe (MapSwipe) module to look at two layers simultaneously. Navigate to the MapSwipe module by clicking on File > Map Swipe (Figure 22).
Figure 22: The path to use GRASS Map Swipe with the g.gui.mapswipe module.
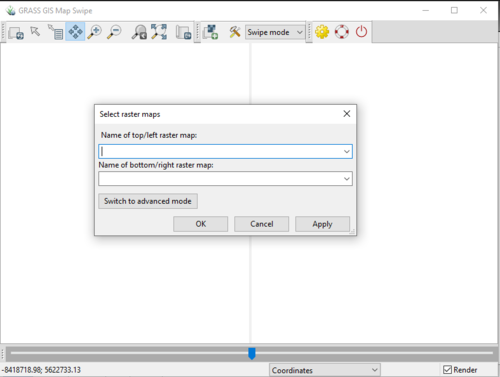
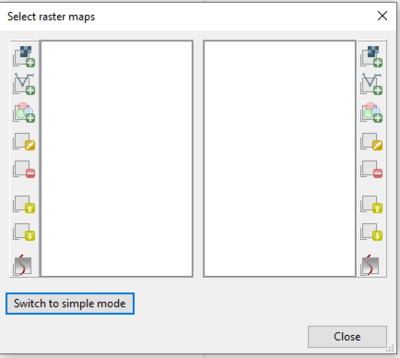
- 6) The MapSwipe module window will open, with another window open on top, the “Select Raster Maps” window (Figure 23). Notice that there is an option to switch to advanced mode, this mode can be seen in Figure 24. Advanced mode gives the user more options for viewing different types of layers. Notice that we could follow the steps above and compare two RGB maps.
Figure 23: The "Select Raster Maps" window from the Map Swipe module.
Figure 24: Advanced mode of the "Select Raster Maps" window showing many different options for selecting layers to compare using the g.gui.mapswipe module.
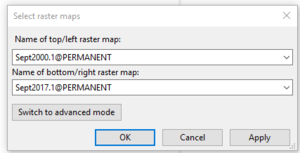
- 7) Switch back to simple mode and select two layers to compare to one another as I have in figure 25. Click OK.
Figure 25: The completed "Select Raster Maps" window.
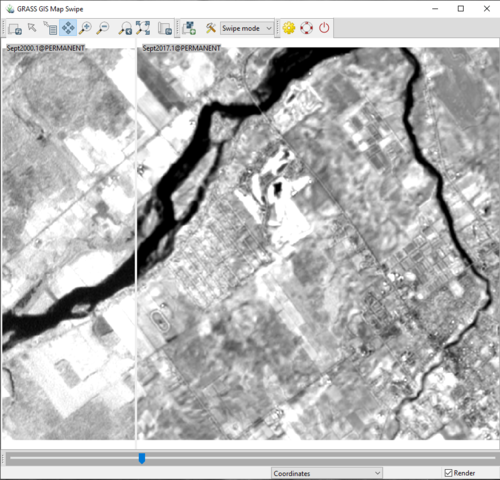
- 8) The “Select Raster Maps” window will close, and you will be able to actively compare the two layers you selected. Click and drag the blue slide at the bottom of the image to see how the two images differ from one another.
Figure 26: The "Map Swipe" window with displaying band 1 of the September 2000 image on the left, and band 1 of the September 2017 image on the right.
- 9) After you are done experimenting with this tool, close the MapSwipe module window to return to the rest of the tutorial.
Raster Smoothing and Discontinuity Maps
The next step in this tutorial is to smooth the raster imagery out and segment the spectral data using the r.smooth.seg module. This module creates two output layers: a smoothed raster map and a discontinuity map. This step is important because smoothing a raster reduces the “noisiness” and makes segmentation more efficient because pixels within a homogeneous area are grouped together and boundaries are smoothed out to reduce the complexity of the segments. This module is also useful because it creates a discontinuity map which contains all of these landscape irregularities so they are not lost after this smoothing step. More information about smoothing, discontinuities and the Mumford-Shah model can be found here and here.
The r.smooth.seg module segments the input raster using an iterative process that can be controlled by altering some of the variables that go into the model. There are two key variables that can be changed in this module to alter the smooth and discontinuity maps: alpha and lambda. Alpha is the discontinuity coefficient which is used to determine the number of allowable discontinuities. Lambda is the smoothness coefficient which is used to determine how smooth the output raster will be. These values must be larger than 0, and if the user does not alter these values manually, alpha and lambda are both provided a value of 1. The authors of this module suggest that users test out different combinations of values to see how their maps change in response to each variable before deciding on values to use in their segmentation process. I carried this process out, but I found that altering alpha and lambda did not change the output dramatically compared to the leaving alpha and lambda at their default values. This is reflected in the steps below, however I strongly suggest that users play with these values because every raster will be different, and this may not always be the case. Finally, the number of iterations that the module carries can be chosen by the user by altering the maximum number of iterations, or by altering the convergence tolerance. In general, the more pixels that the map contains, the more iterations it will take to segment it. More information about the r.smooth.seg module can be found here.
- 1) Navigate to the “Modules” tab on the “Layer Manager” window. Expand the “Addons” category, and double-click the r.smooth.seg module.
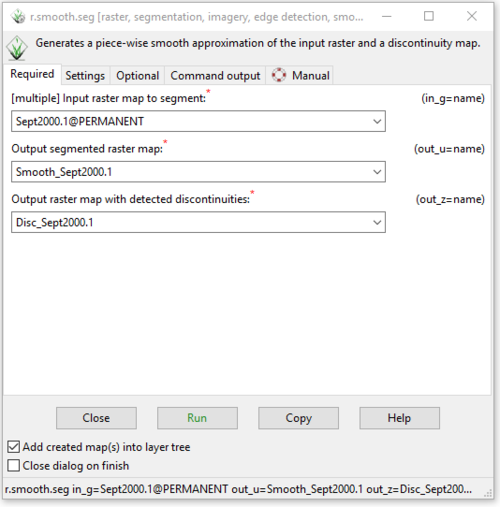
- 2) Complete the information on the “Required” tab in the r.smooth.seg module window as seen in Figure 27.
Figure 27: The "r.smooth.seg Module" window with the completed input and output information on the "Required" tab.
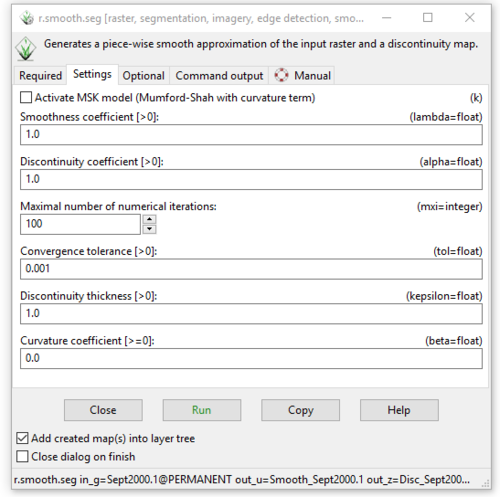
- 3) Click on the “Settings” tab to see the other variables this module has. As discussed above, leave these variables at their default settings see in Figure 28.
- 4) Click Run. Repeat these steps for the other bands from the Sept2000 and Sept2017 layers.
Figure 28: The "Settings" tab in the "r.smooth.seg Module" window showing the different variables that can be adjusted.
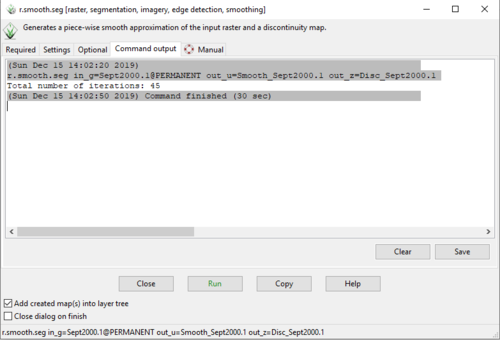
- 5) Navigate to the “Command Output” tab. You will see that for this band, 45 iterations were needed to smooth the imagery out until it reached a convergence tolerance of 98% (Figure 29). In general, if the layer requires all 100 iterations (default value) to complete the smoothing process, you may increase the number of allowable iterations, or alter the values of alpha, lambda, or the convergence tolerance.
Figure 29: The "Command Output" tab in the "r.smooth.seg Module" window showing the output information.
- 6) When all of the bands have been input to this module, you may close the r.smooth.seg module window.
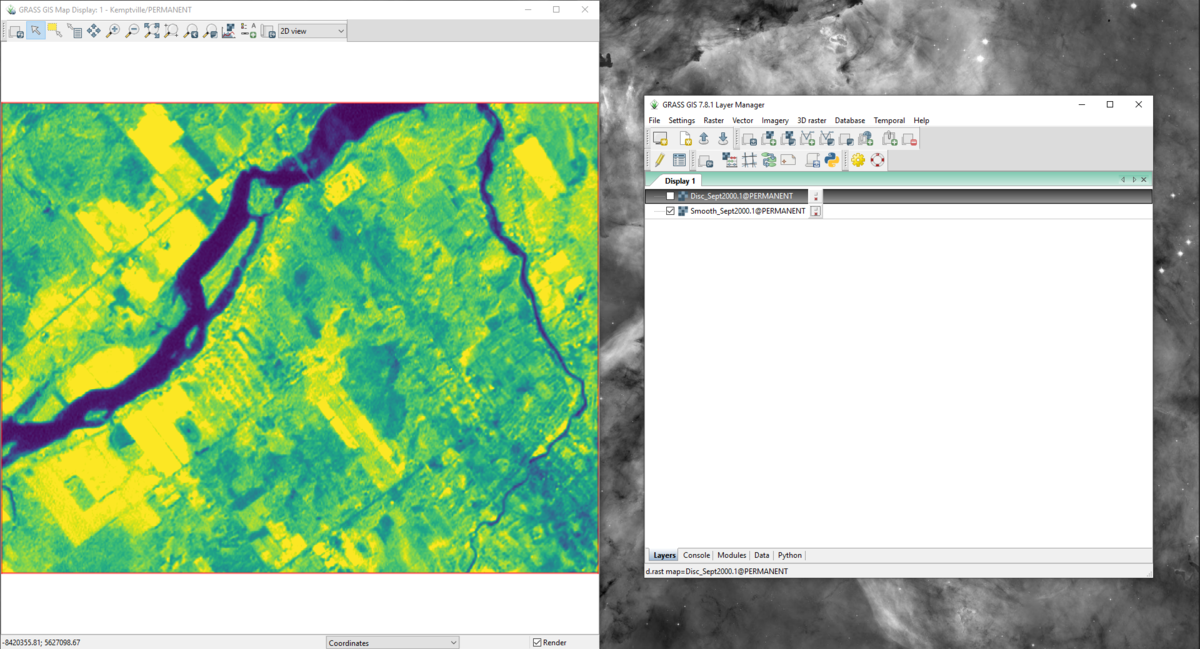
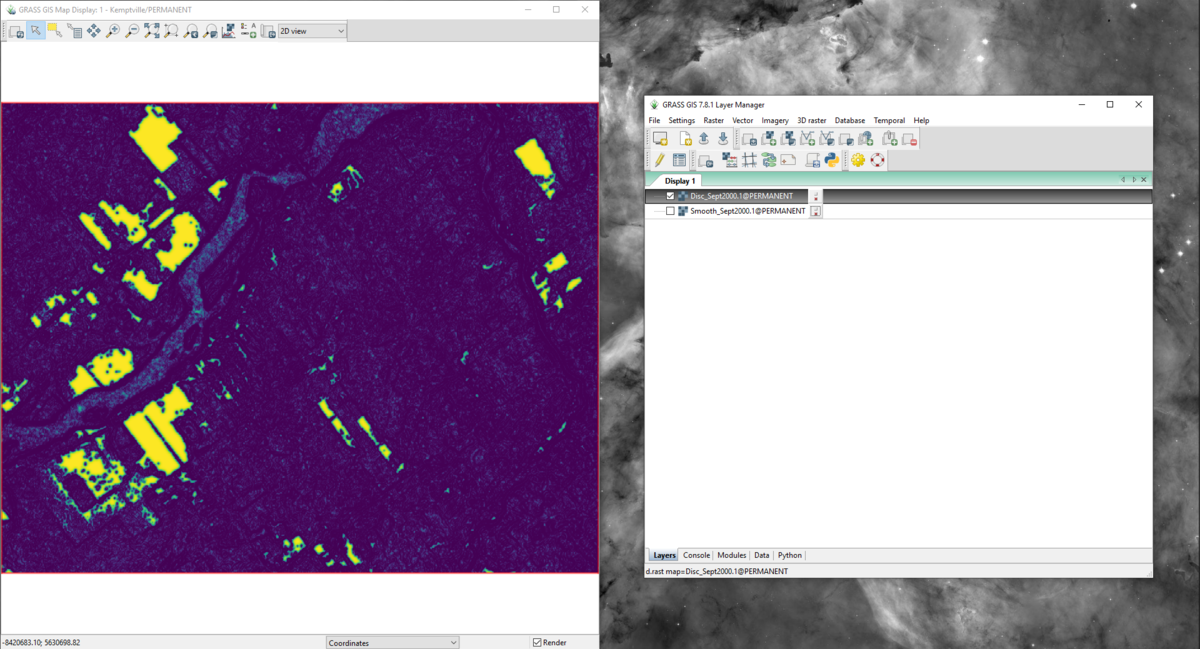
- 7) You may look at the output layers created for each band by selecting the layers in the “Layer Manager” window so they appear in the “Map Display” window. Figure 30 and 31 provide an example of the smoothed and discontinuity maps for band 4 of the September 2000 data (Sept2000.1).
Figure 30: The "Map Display" window with the smoothed map output for band 4 of the September 2000 data using the r.seg.smooth module.
Figure 31: The "Map Display" window with the discontinuity map output for band 4 of the September 2000 data using the r.seg.smooth module.
Image Grouping
The next step is to put our band layers into groups based on year. Here we are recombining bands 2, 3, and 4 for September 2000 and September 2017 into their respective groups using the i.group module. This tool allows the user to run analyses on combinations of data. Many imagery analysis modules in GRASS require the user to group their data, and some modules even require the user to create further subgroups within these groups. The i.cluster and i.maxlik modules are used later in the tutorial and are examples of modules that require subgroups. In this tutorial, the group and subgroups will contain the exact same imagery, however users may group and subgroup their data based off of different criteria and this may not always be the case. Users may also come back to this tool to edit a group/subgroup after it is created. More information about the i.group module can be found here.
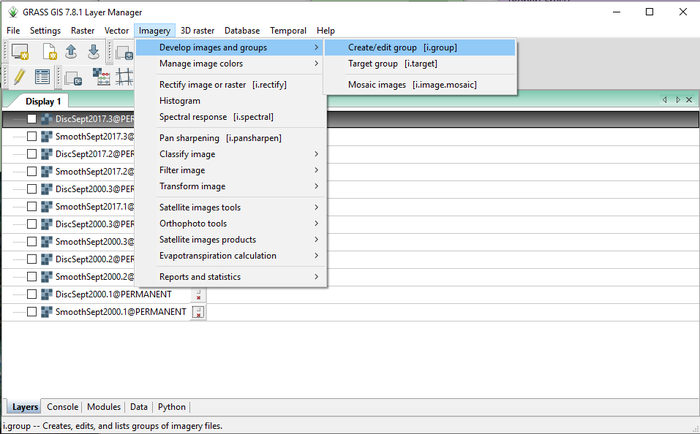
- 1) Navigate to the i.group module by clicking on Imagery > Develop Images and Groups > Create/Edit Group (Figure 32).
Figure 32: The path to the i.group module in the "Layer Manager" window.
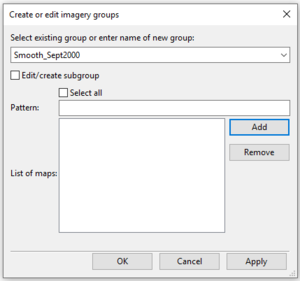
- 2) Type a name for your group in the “Enter name of a new group:” box in the i.group module window (Figure 33). If you were editing an existing group, you would click on this box to expand it and select the group you wish to edit.
Figure 33: The "Create or Edit Imagery Groups" window from the i.group module.
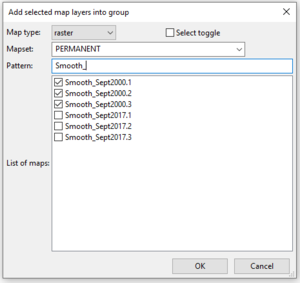
- 3) Click Add to select the layers you would like to group together. The “Add selected map layers into group” window will open (Figure 34).
- 4) Begin typing the name of the layer you would like to add into your group and select the layers from the “List of maps” section.
Figure 34: The "Add Selected Layers into Group" window from the i.group module.
- 5) Click OK, then click Apply in the i.group module window. This creates the group with all of the photos you have selected.
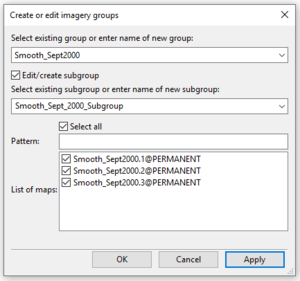
- 6) Tick the box beside "Edit/Create Subgroup".
- 7) Type the name of the subgroup into the “Enter name of a new subgroup:” box. If you were editing an existing subgroup, you would click on this box to expand it and select the subgroup you wish to edit.
- 8) Tick the box beside “Pattern – Select all” (Figure 35). You may also select the images from the “List of maps” that you would like to include in this subgroup.
Figure 35: The "Create Imagery Groups" window showing the creation of a subgroup using the i.group module.
- 9) Click Apply then click OK. The i.group module window will close.
Category Clustering
Next, we will apply a clustering algorithm to both subgroups of images we created in the previous step. The i.cluster module uses the spectral reflectance of each pixel in the imagery data to cluster the pixels into categories that can then be related to different land-use types. The algorithm that clusters the pixels is a modified version of the k-means clustering algorithm. The spectral signatures of classes in the image are compiled into an output file that is used by the i.maxlik module for an unsupervised classification of the land-use types. The user determines how many classes they would like to categorize the pixels into.
An important factor to mention is that the i.cluster module does not cluster all of the pixels in the image, rather it clusters approximately 10 000 pixels and creates signatures for the file based on the clusters. Users have the ability to change the sampling pattern by updating the sample parameter to sample every x-th column, and every y-th column. The smaller these x- and y- values are, the more pixels that are sampled. Apart from the sampling method, there are several variables that may be altered that impact the distribution of the clusters.
The minimum number of pixels, the number of allowable iterations, and the convergence percent are all very important for this module. The minimum number of pixels is set to 17 pixels by default. This parameter is the minimum number of pixels that make up a class. As you increase the number of classes you would like to differentiate between, this number matter more. Because this tutorial is only going to classify our image in to 4 categories of land-use, this value is not changed.
The convergence percent and the number of allowable iterations are the parameters which determine when the clustering algorithm stops clustering pixels. The convergence percent refers to the point at which the means of the clusters become stable. This is the primary force that causes the clustering algorithm to stop. In general, if the number of iterations reached is the maximum number of allowable iterations, it can be assumed that the convergence percent was not reached. If this is the case, the user can alter these two values to either cause the module to stop processing the cluster means at a lower convergence point, or to increase the number of iterations that occur. The convergence percent and number of allowable iterations are set to 98% and 30 iterations by default. More information about the i.cluster module can be found here.
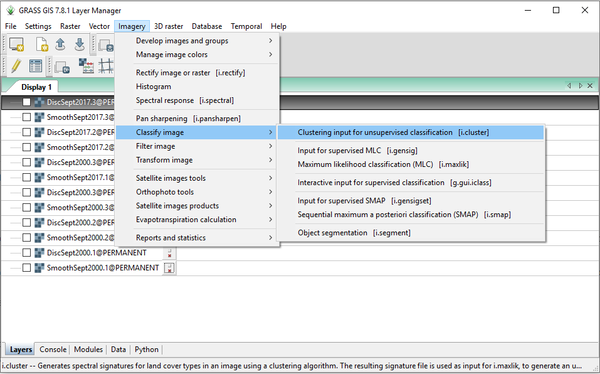
- 1) Navigate to the i.cluster module by clicking on Imagery > Classify Image > Clustering Input for Unsupervised Classification (Figure 36).
Figure 36: The path to the i.cluster module in the "Layer Manager" window.
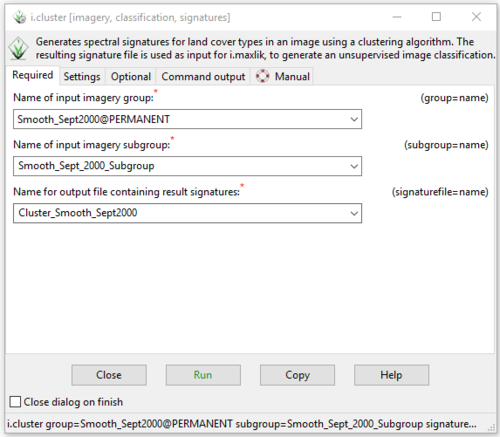
- 2) The i.cluster module window will open to the “Required” tab. Input the group and subgroup for one of the images we created in the previous step, and then type in a name for the output signature file that the i.cluster module will produce (Figure 37).
Figure 37: The "i.cluster Module" window showing the input and output information on the Required tab
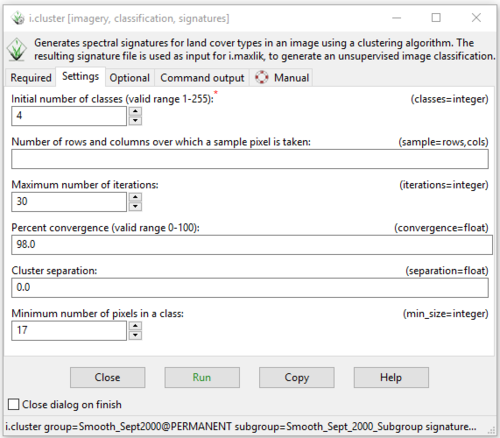
- 3) On the “Settings” tab, the other parameters mentioned can be altered. The user must enter the number of classes that they would like the clustering algorithm to produce, in this case, type “4” into the “Initial number of classes:” box (Figure 38).
- 4) Click Run, then repeat these steps for the other group/subgroup you created.
Figure 38: The "i.cluster Module" window showing the variables available on the Settings tab.
Unsupervised Classification
In this section, we will be using the i.maxlik module to classify the spectral reflectance of our images using the output signature information file we just generated in the previous step with the i.cluster module. This tool is a maximum-likelihood discriminant analysis classifier and is used in both unsupervised image classification (what we are doing here) and supervised image classification. This module works by taking each cell in the image and determining which cluster it is most likely to belong in based on the cluster means and covariance matrices produced by the i.cluster module. This module produces a classified image where each cell belongs to a specific spectral class, which can then be related to a particular land-use type. The module also has the ability to produce a raster that contains the reject threshold results. This output is useful for determining which cells/areas have a low probability of being assigned to the right class. More information about the i.maxlik module can be found here.
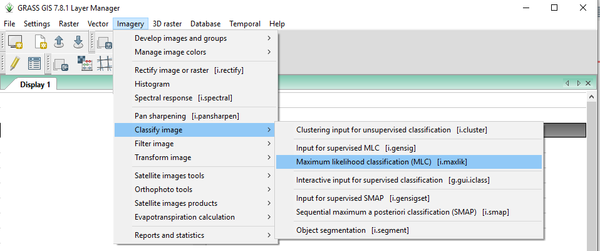
- 1) Navigate to the i.maxlik module by clicking on Imagery > Classify Image > Maximum Likelihood Classification (MLC) (Figure 39).
Figure 39: The path to the i.maxlik module in the "Layer Manager" window.
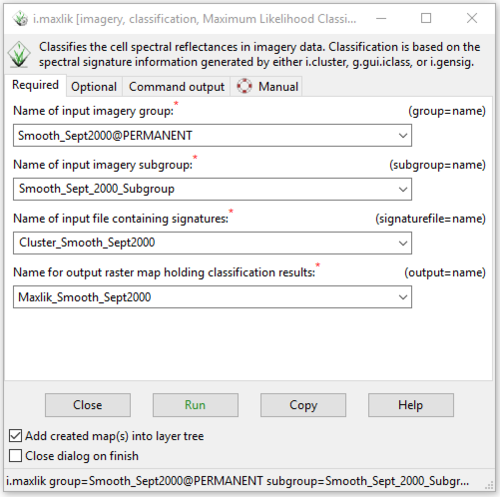
- 2) The i.maxlik module window will open to the “Required” tab. Input the group and subgroup for one of the images, and then select the signature file that was produced in the previous section. Finally, enter a name for the output raster in the “Name for output raster map holding classification results:” box (Figure 40).
Figure 40: The "i.maxlik Module" window showing the input and output information on the Required tab
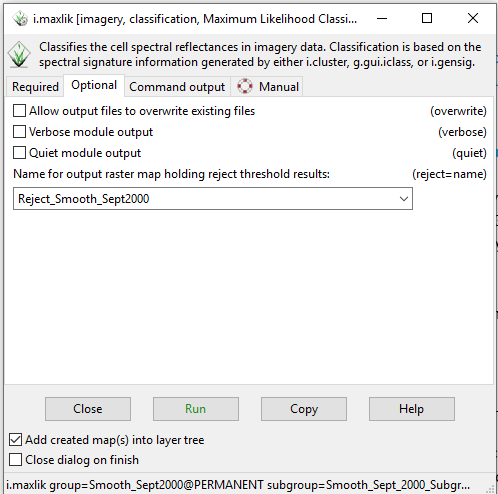
- 3) Click on the “Optional” tab and type a name for the reject output raster into the “Name for output raster map holding reject threshold results:” box (Figure 41).
Figure 41: The "i.maxlik Module" window showing the input and output information on the Optional tab
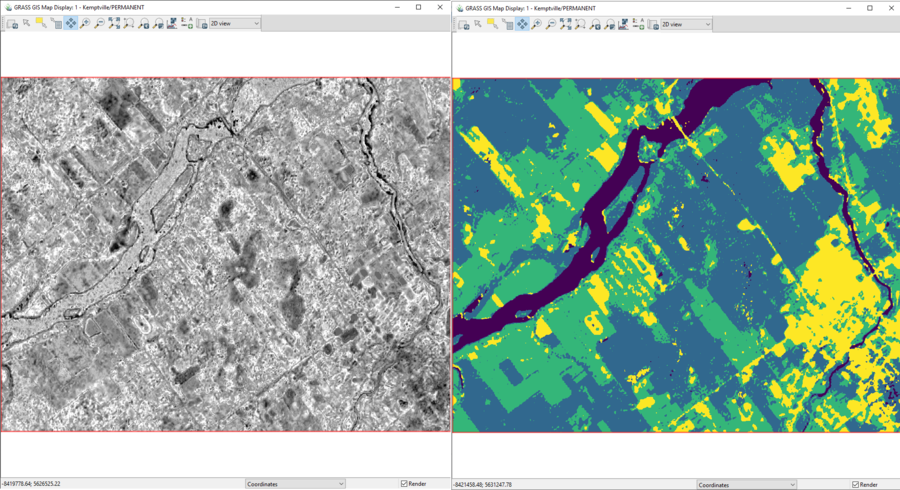
- 4) Click Run to complete this step. The classified map and the reject threshold raster should look similar to the ones in Figure 42.
Figure 42: The classified map (right) and the reject threshold output raster (left) for September 2000 created by the i.maxlik module.
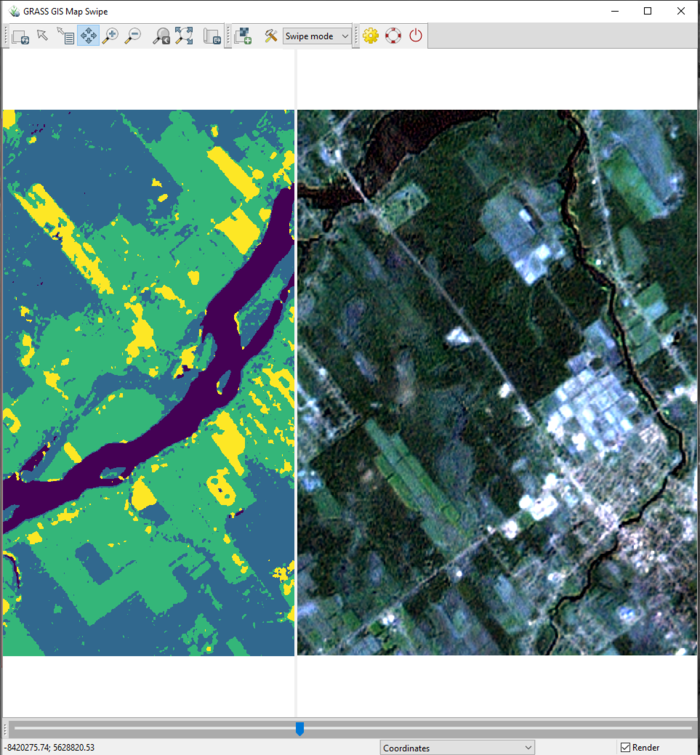
- 5) To estimate which classified colour represents which land-use type, I suggest using the MapSwipe tool to compare the classified images with the visible band imagery provided (Visible_7Sept2000.tiff and Visible_23Sept2017.tiff). This comparison is seen in Figure 43. From this information, we can see that in the classified image, purple represents water, blue represents dense vegetation (forest), green represents thinner vegetation (likely agricultural land), and yellow represents developed land (houses, roads).
Figure 43: The Map Swipe window showing the classified imagery for September 2000 on the left, and the visible bands (1, 2, and 3) for September 2000 on the right.
Landscape Change Assessment
The final step in this tutorial is to compare the land-use types in September 2000 and September 2017. To do this, we will use the r.change.info module we downloaded into GRASS earlier in the tutorial. This module calculates the change in landscape according to different measures of landscape composition (land-use types), configuration (arrangement of land-use types), or a combination of both composition and configuration. There are six methods of change assessment that can be calculated with this module: proportion of change, information gain, information gain ratio, Chi-square, Gini impurity, and distance (referring to the statistical distance between the average and observed distributions). Depending on the research question you have, you may use different methods of change assessment. In this tutorial, I will be showing you how to measure the proportion of change and information gain. With all of these methods of change assessment, a larger value means a larger difference between the two input maps. This module also provides the user the option of selecting one measure of change, or multiple methods of change assessment to calculate between the two input rasters. In this section, I will provide instructions for both methods.
Aside from the different methods of landscape change assessment, there are three parameters which will alter the result of these tests. The window size, processing step, and alpha for general entropy. The window size refers to the size (in cells) of the processing window. By altering the window size, you change the extent over which the module processes the two input maps. The processing step refers to the resolution over which the changes are identified. As a result, the step cannot be larger than the size. When the step is decreased, the resolution increases. Both the window size and processing step have a default value of 40 cells. Finally, information gain and its ratio include a measure of entropy. By default, Shannon’s entropy is the measure that is used. This measure of entropy can be changed to reflect different measures of Renyi’s entropy which changes the weight of changes, and can be very useful for noisy data that may not be classified accurately. More information about the r.change.info module and the different methods of change assessment can be found here.
- 1) Navigate to the “Modules” tab on the “Layer Manager” window. Expand the “Addons” category, and double-click the r.change.info module.
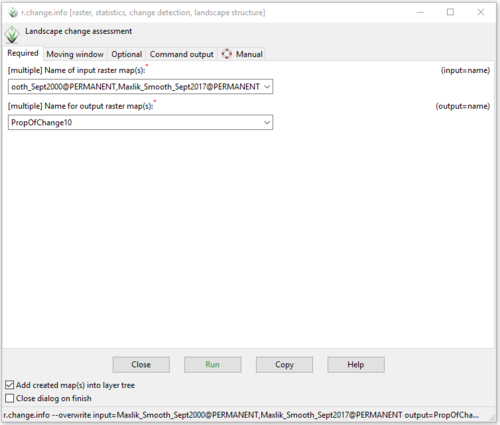
- 2) First, I will show you how to calculate one method of change assessment. Complete the information on the “Required” tab in the r.smooth.seg module window as seen in Figure 44. Notice that both classified raster layers are included as input raster maps in this module.
Figure 44: The "r.change.info Module" window with the input and output information on the "Required" tab for the "PropOfChange10" layer.
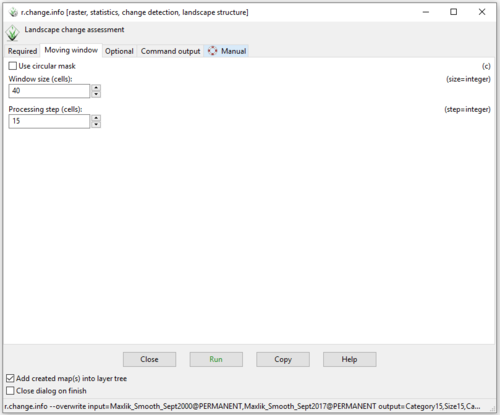
- 3) Click on the “Moving Window” tab, and change the processing step to 10 cells (Figure 45). I found that a processing step of 10 or 15 cells worked very well for this data set, however feel free to experiment with this value.
Figure 45: The "r.change.info Module" window with variable information on the "Moving Window" tab, where step = 10 cells.
- 4) Click on the “Optional” tab and tick the box for the “Proportion of Changes” method of change assessment (Figure 46).
Figure 46: The "r.change.info Module" window with the "proportion of changes" change assessment selected on the "Optional" tab.
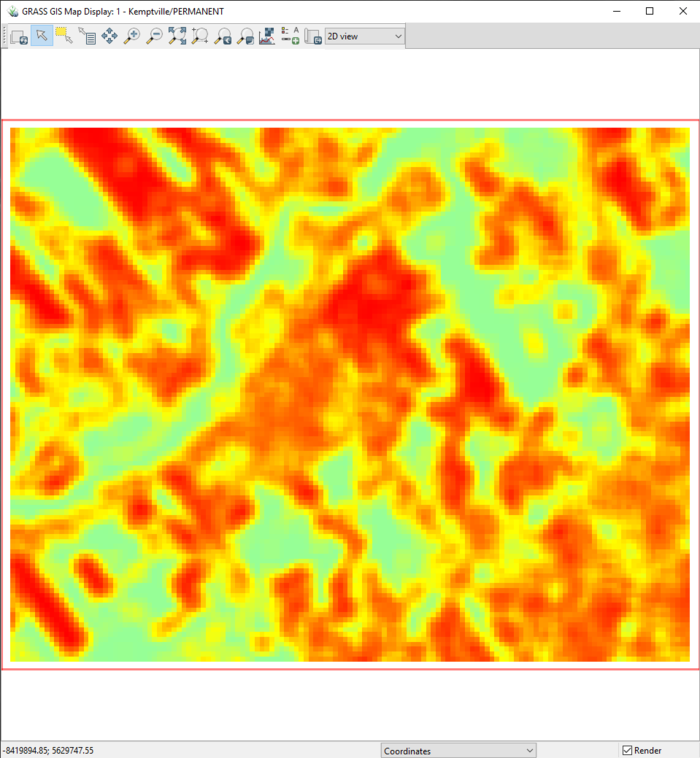
- 5) Click Run. The proportion of change output raster should look like the one in Figure 47. Notice the resolution here as you can compare it to the resolution used in the next steps.
Figure 47: The output from the r.change.info module with the "PropOfChange40" layer in the "Map Display" window.
- 6) Next, I will show you how to calculate multiple methods of change assessment in one step. Moving back to the “Required” tab, this time you will type in multiple names for the output rasters, separated by commas (Figure 48). Again, notice that both classified raster layers are included as input raster maps.
Figure 48: The "r.change.info Module window" with the input and output information on the "Required" tab for the "Category15","Size15", and "Category_Size15" layers.
- 7) Click on the “Moving Window” tab. This time, we will run the module with a step size of 15 cells (Figure 49) to compare to the step size of 10 cells seen above in Figure 45.
Figure 49: The "r.change.info Module" window with variable information on the "Moving Window" tab, where step = 15 cells.
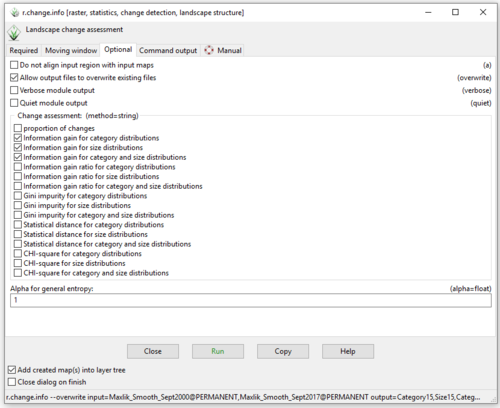
- 8) Click on the “Optional” tab and this time, tick the box beside multiple methods of change assessment (Figure 50). Make sure that the order of the output raster names you chose on the “Required” tab matches the order in which they appear in the “method of change assessment” list on the “Optional” tab and not the order in which you selected the methods.
Figure 50: The "r.change.info Module" window with "Information Gain for Category Distributions", "Information Gain for Size Distributions", and "Information Gain for Category and Size Distributions" change assessments selected on the Optional tab.
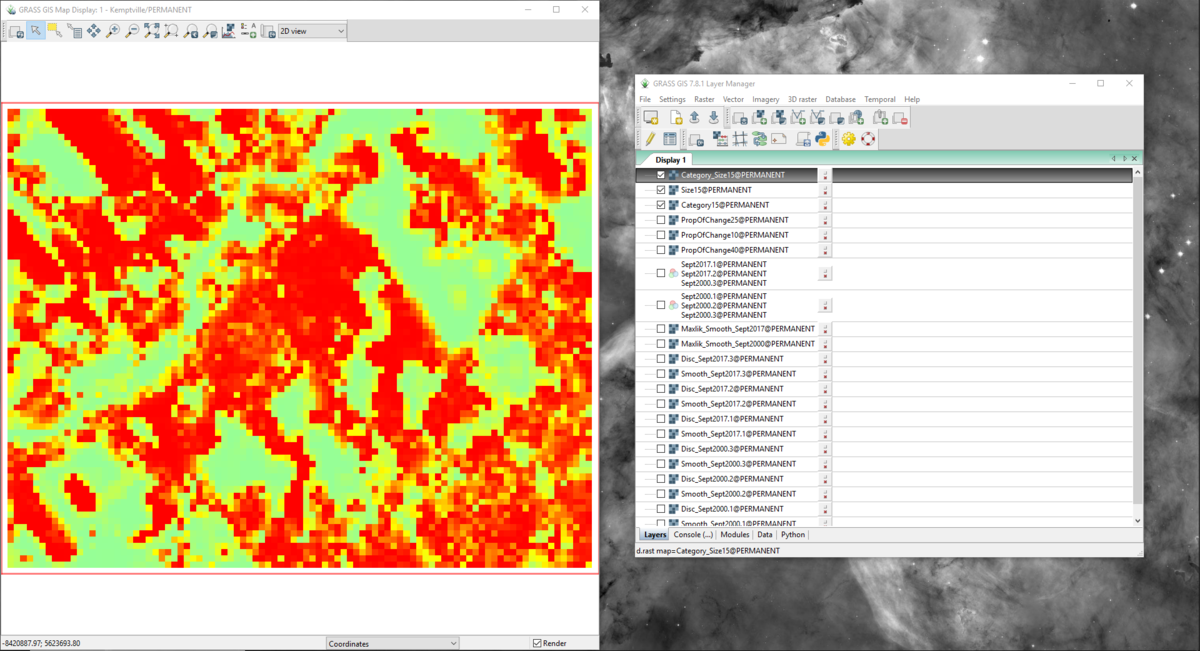
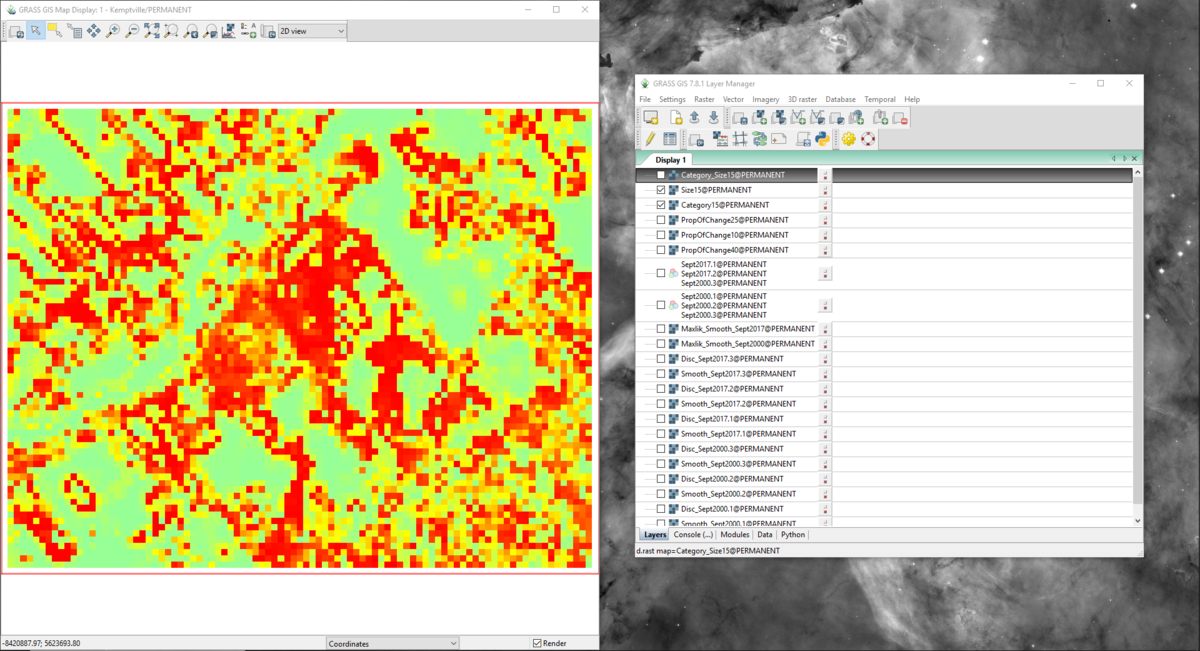
- 9) Click Run. This time there will be 3 new output rasters created, and you can look at them individually by selecting them on the “Layer Manager” window. The output rasters should look like the ones that appear in figures 51, 52, and 53. Notice that these layers vary from one another visually. The output of this tool creates cells on a range of high to low change, where the colour red represents large changes in landscape and green represents small or no changes. The next section will focus on adding a legend to the map layout to determine how big the changes in landscape are.
Figure 51: The output from the r.change.info module with the "Category_Size15" layer in the "Map Display" window.
Figure 52: The output from the r.change.info module with the "Size15" layer in the "Map Display" window.
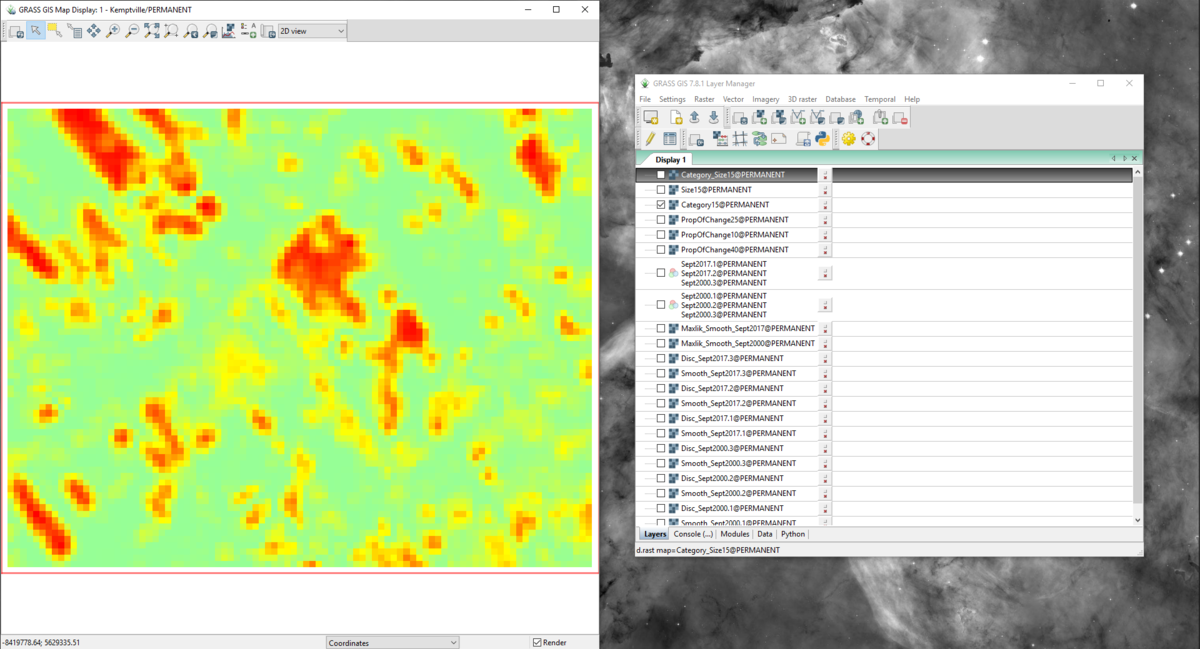
Figure 53: The output from the r.change.info module with the "Category15" layer in the "Map Display" window.
Adding a Legend
In this last section, we will be looking at adding a legend to our landscape change maps to allow for further interpretation. There are many different options that can be adjusted using the d.legend module, including the addition of titles, alteration of fonts, font sizes, font colours, and even the addition of histograms. We will use this module to add a smooth gradient legend to our map, however there is no limit on what you can do to improve the clarity of your map. More in formation about the d.legend module can be found here.
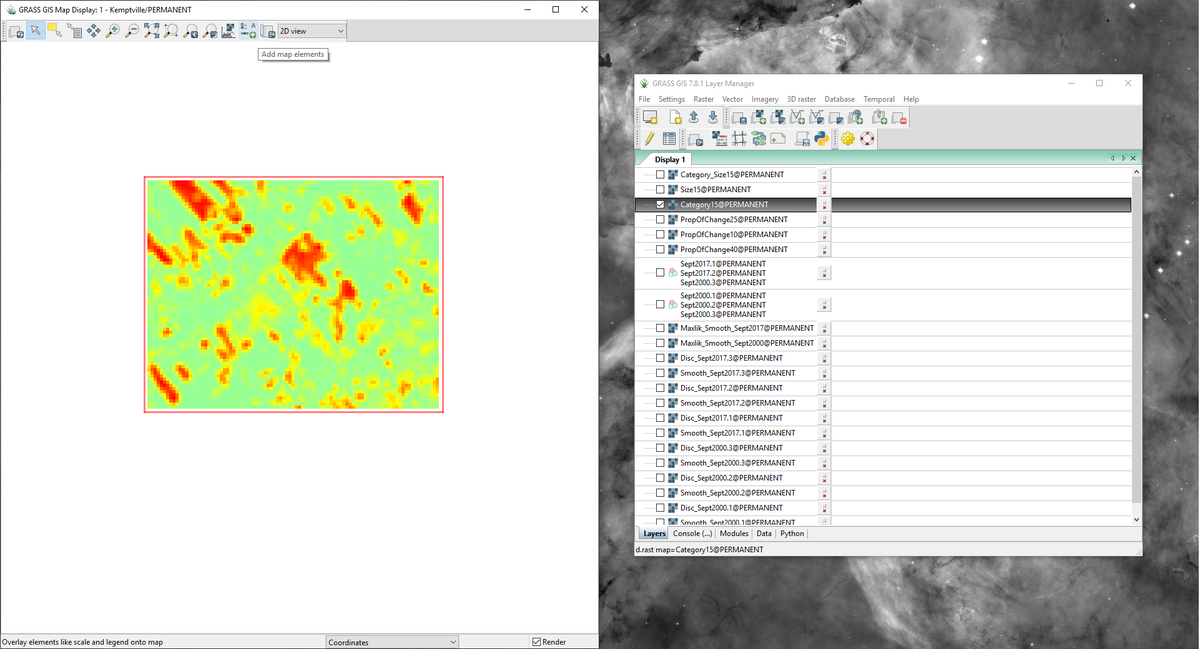
- 1) Make sure that you have selected the layer you would like to alter in the “Layer Manager” window, and that the correct layer appears in the “Map Display” window. I suggest that you zoom out a bit to provide some whitespace on the layout (Figure 54).
Figure 54: The "Category15" layer in the "Map Display" window zoomed out.
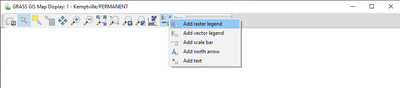
- 2) Navigate to the d.legend module in the “Map Display” window by following the path seen in Figure 555.
Figure 55: The path to the "Add Raster Legend" module (d.legend) in the "Map Display" window.
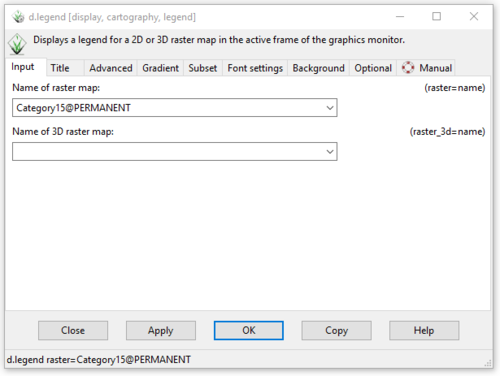
- 3) The d.legend module window will open to the “Input” tab. Ensure that the name of the raster map is indeed the one you would like to adjust (Figure 56).
Figure 56: The "Input" tab in the "d.legend Module" window.
- 4) Switch to the “Gradient” tab and check the box that says, “Draw Smooth Gradient” (Figure 57). Feel free to explore the many different options that module offers that you can take advantage of in the other tabs.
Figure 57: The "Gradient" tab in the "d.legend Module" window showing where the labels for the gradient legend may be altered.
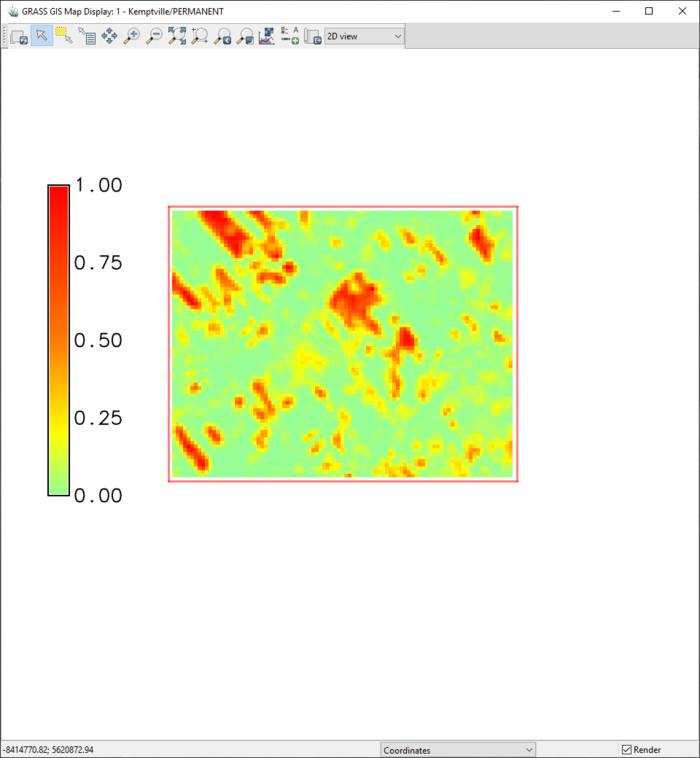
- 5) Click Apply, then OK, then Close. The smooth gradient will now appear on the layout (Figure 58). It can be moved easily by clicking and dragging the object. The alter the object, double-click on it to re-launch the d.legend module. To delete the object, right click on it and select Remove Legend.
Figure 58: The "Category15" layer with a gradient legend.
Conclusion
This tutorial provides users with freely available satellite imagery, open-source software, and instructions on how to conduct unsupervised classification with a temporal analysis of land-use change. Users are encouraged to explore GRASS GIS further and work with other spatio-temporal modules especially the r.li toolset. More information about this toolset can be found here.
The resulting output maps in this tutorial show that the area of interest in Kemptville, Ontario showed many changes to land-use between 2000 and 2017 according to the proportion of change and information gain methods of change assessment used. At a glance, it appears that there were greater changes to landscape configuration (arrangement of land-use types, size) compared to composition (land-use type, category) as seen in Figures 51 and 52. However, these changes appear to be dramatically enhanced when the two measures of landscape are combined (Figure 50). In conclusion, I hope that users will take these steps and apply them to their own research questions. I also hope that users will attempt to conduct statistical analyses of the changes in land-use types which will allow them to determine which measures of landscape have changed the most over a given time period.
References
- [1] NASA's Landsat Science Homepage
- [2] GRASS GIS Homepage
- [3] OSGeo Foundation Homepage
- [4] Introduction to GRASS Workshop (Tutorial)
- [5] Earth Observing System (EOS) Landviewer
- [6] Shaw, M. (2017) Building boom in Kemptville, Ont., to continue … for years. Real Estate News Exchange.
- [7] GRASS GIS User Manual Homepage
- [8] GRASS GIS Quickstart Guide
- [9] GRASS GIS Addons Homepage
- [10] GRASS GIS Manual - r.in.gdal Module
- [11] GDAL Library Homepage
- [12] GRASS GIS Manual - d.rgb Module
- [13] GRASS GIS Manual - g.gui.mapswipe Module
- [14] Vitti, A. (2008) Free Discontinuity Problems in Image and Signal Segmentation. PhD Thesis. University of Trento. 1-124.
- [15] Vitti, A. (2012) The Mumford–Shah variational model for image segmentation: An overview of the theory, implementation and use. ISPRS Journal of Photogrammetry and Remote Sensing. 69: 50-64.
- [16] GRASS GIS Manual - r.smooth.seg Module
- [17] GRASS GIS Manual - i.group Module
- [18] GRASS GIS Manual - i.cluster Module
- [19] GRASS GIS Manual - i.maxlik Module
- [20] GRASS GIS Manual - r.change.info Module
- [21] GRASS GIS Manual - d.legend Module
- [22] GRASS GIS Manual - r.li Toolset