Modelling Air Mass Time Over Water with Hysplit, SAGA GIS, and Python
Contents
Introduction
In this tutorial, we will explore how to combine a vector line with raster surface information to extract quantitative information about the conditions underneath the line. There are numerous possible applications for this: we can look at an air mass moving over different land cover types, the impact of pollutants on river currents, or the habitats an animal prefers to travel through. Here, we will focus on one particular application, that of how long an air mass spends over water. However, the techniques can be applied to many different problems.
For our purposes, the line will represent the trajectory of an air mass, with each segment representing a certain amount of time. We are interested in how long the air mass spent over water. This is of interest since air masses that spend a long time over water tend to have higher humidity which can affect how much evaporation occurs at further points.
Given a raster grid that determines whether a cell is water or not, we can evaluate this by the following general process:
- Split the line into segments based on the raster grid
- Determine whether or not the segment is over water and how much time it represents
- Sum the length of time represented by segments that are over water.
We will also briefly demonstrate how to use a National Oceanic and Atmospheric Administration (NOAA) tool called Hysplit to determine the backtrajectory of an air mass from a point in time and space over a week. Then, we'll see how Python can be used to extract that data and, using SAGA GIS, calculate the amount of time spent over water for the trajectory. The process can be adapted with minimal changes to use other trajectories and raster data.
Prerequisites
- Thorough comfort with Python and command line tools would be greatly beneficial. You might find Automating SAGA Workflows Using Command Line Scripting useful to start. A course in Python programming (such as COMP 1005 here at Carleton) is a great foundation but the tools introduced here will go beyond it.
- Comfort with your operating system is recommended.
- The optional Hysplit component requires some comfort with FTP servers.
This tutorial is focused on the Windows environment, but all the software described should also work in a Unix environment with suitable modifications.
Setup
Hysplit
An example Hysplit data file is provided below if you’d rather not follow these steps. However, if you are interested in air mass trajectory analysis, you may find these useful.
- Visit the Hysplit utilities page and install the Tcl/Tk graphical user interface and the Ghostscript / Ghostview postscript viewer. Make sure you install Ghostscript and GSview to their default directories to avoid complications.
- You can then download and install Hysplit
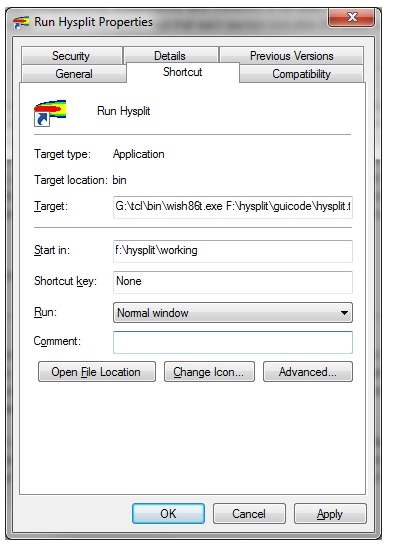
- To run Hysplit on Windows, you’ll need to edit the shortcut to tell it to run the Hysplit Tcl code with the Tcl interpreter. Right click on the shortcut to Hysplit on your desktop or start menu and click Properties. Edit the “Target” field to have the path to the Tcl executable called wish86t.exe. On other platforms, you may have to take similar steps.
- You can then test that you can launch Hysplit.
Otherwise, copy and paste the following content into a text file and save it as a file named "tdump".
1 1
NARR 19 12 1 0 0
1 BACKWARD OMEGA
19 12 21 21 45.388 -75.696 2.0
1 PRESSURE
1 1 19 12 21 21 0 0 0.0 45.388 -75.696 2.0 1013.9
1 1 19 12 21 20 0 0 -1.0 45.337 -75.589 0.0 1015.5
1 1 19 12 21 19 0 0 -2.0 45.301 -75.462 0.0 1017.6
1 1 19 12 21 18 0 0 -3.0 45.282 -75.314 0.0 1020.4
1 1 19 12 21 17 0 0 -4.0 45.278 -75.159 0.0 1022.4
1 1 19 12 21 16 0 0 -5.0 45.284 -75.014 0.0 1023.2
1 1 19 12 21 15 0 0 -6.0 45.296 -74.885 0.0 1024.4
1 1 19 12 21 14 0 0 -7.0 45.306 -74.769 0.0 1025.2
1 1 19 12 21 13 0 0 -8.0 45.310 -74.661 0.0 1025.9
1 1 19 12 21 12 0 0 -9.0 45.310 -74.557 0.0 1026.9
1 1 19 12 21 11 0 0 -10.0 45.312 -74.460 0.0 1028.0
1 1 19 12 21 10 0 0 -11.0 45.317 -74.373 0.0 1029.6
1 1 19 12 21 9 0 0 -12.0 45.322 -74.301 0.2 1030.4
1 1 19 12 21 8 0 0 -13.0 45.324 -74.251 0.4 1030.0
1 1 19 12 21 7 0 0 -14.0 45.322 -74.231 0.4 1029.9
1 1 19 12 21 6 0 0 -15.0 45.316 -74.239 0.2 1030.1
1 1 19 12 21 5 0 0 -16.0 45.306 -74.270 0.0 1030.4
1 1 19 12 21 4 0 0 -17.0 45.292 -74.320 0.0 1030.4
1 1 19 12 21 3 0 0 -18.0 45.275 -74.385 0.0 1029.7
1 1 19 12 21 2 0 0 -19.0 45.260 -74.458 0.0 1029.7
1 1 19 12 21 1 0 0 -20.0 45.253 -74.534 0.0 1029.9
1 1 19 12 21 0 0 0 -21.0 45.254 -74.615 0.0 1030.4
1 1 19 12 20 23 0 0 -22.0 45.256 -74.700 0.0 1029.7
1 1 19 12 20 22 0 0 -23.0 45.254 -74.788 0.0 1029.3
1 1 19 12 20 21 0 0 -24.0 45.244 -74.883 0.0 1028.9
1 1 19 12 20 20 0 0 -25.0 45.227 -74.997 0.0 1029.9
1 1 19 12 20 19 0 0 -26.0 45.200 -75.142 0.0 1030.4
1 1 19 12 20 18 0 0 -27.0 45.166 -75.314 0.0 1030.9
1 1 19 12 20 17 0 0 -28.0 45.144 -75.490 0.0 1028.1
1 1 19 12 20 16 0 0 -29.0 45.153 -75.651 0.6 1025.4
1 1 19 12 20 15 0 0 -30.0 45.202 -75.802 2.4 1023.3
1 1 19 12 20 14 0 0 -31.0 45.271 -75.953 5.0 1023.0
1 1 19 12 20 13 0 0 -32.0 45.335 -76.106 7.3 1022.1
1 1 19 12 20 12 0 0 -33.0 45.398 -76.258 8.8 1017.9
1 1 19 12 20 11 0 0 -34.0 45.471 -76.410 9.6 1013.1
1 1 19 12 20 10 0 0 -35.0 45.562 -76.562 9.7 1010.0
1 1 19 12 20 9 0 0 -36.0 45.659 -76.716 9.3 1008.6
1 1 19 12 20 8 0 0 -37.0 45.740 -76.868 8.1 1008.7
1 1 19 12 20 7 0 0 -38.0 45.805 -77.010 5.2 1009.4
1 1 19 12 20 6 0 0 -39.0 45.862 -77.142 1.1 1010.3
1 1 19 12 20 5 0 0 -40.0 45.916 -77.270 0.0 1009.3
1 1 19 12 20 4 0 0 -41.0 45.967 -77.396 2.9 1008.1
1 1 19 12 20 3 0 0 -42.0 46.015 -77.520 11.8 1006.9
1 1 19 12 20 2 0 0 -43.0 46.063 -77.634 26.0 1000.6
1 1 19 12 20 1 0 0 -44.0 46.109 -77.743 32.5 995.7
1 1 19 12 20 0 0 0 -45.0 46.147 -77.856 28.0 993.4
1 1 19 12 19 23 0 0 -46.0 46.177 -77.963 21.3 992.5
1 1 19 12 19 22 0 0 -47.0 46.207 -78.052 15.8 991.9
1 1 19 12 19 21 0 0 -48.0 46.239 -78.124 11.6 991.3
1 1 19 12 19 20 0 0 -49.0 46.269 -78.198 8.6 991.3
1 1 19 12 19 19 0 0 -50.0 46.289 -78.285 6.5 991.1
1 1 19 12 19 18 0 0 -51.0 46.297 -78.385 5.1 990.9
1 1 19 12 19 17 0 0 -52.0 46.295 -78.488 4.2 990.6
1 1 19 12 19 16 0 0 -53.0 46.288 -78.587 3.4 990.4
1 1 19 12 19 15 0 0 -54.0 46.279 -78.682 3.0 989.6
1 1 19 12 19 14 0 0 -55.0 46.273 -78.778 3.3 989.4
1 1 19 12 19 13 0 0 -56.0 46.277 -78.874 4.7 989.5
1 1 19 12 19 12 0 0 -57.0 46.290 -78.968 7.1 989.9
1 1 19 12 19 11 0 0 -58.0 46.309 -79.064 9.0 989.8
1 1 19 12 19 10 0 0 -59.0 46.332 -79.160 9.1 990.2
1 1 19 12 19 9 0 0 -60.0 46.359 -79.268 6.9 991.9
1 1 19 12 19 8 0 0 -61.0 46.391 -79.393 3.5 992.9
1 1 19 12 19 7 0 0 -62.0 46.421 -79.532 0.9 994.4
1 1 19 12 19 6 0 0 -63.0 46.450 -79.678 0.3 995.2
1 1 19 12 19 5 0 0 -64.0 46.494 -79.818 2.2 995.6
1 1 19 12 19 4 0 0 -65.0 46.564 -79.951 5.9 994.8
1 1 19 12 19 3 0 0 -66.0 46.650 -80.074 9.6 991.0
1 1 19 12 19 2 0 0 -67.0 46.755 -80.203 12.8 987.5
1 1 19 12 19 1 0 0 -68.0 46.894 -80.355 17.5 984.1
1 1 19 12 19 0 0 0 -69.0 47.081 -80.546 20.2 978.1
1 1 19 12 18 23 0 0 -70.0 47.300 -80.764 19.2 970.4
1 1 19 12 18 22 0 0 -71.0 47.507 -80.987 17.3 969.6
1 1 19 12 18 21 0 0 -72.0 47.699 -81.193 17.0 969.0
1 1 19 12 18 20 0 0 -73.0 47.894 -81.404 20.4 968.9
1 1 19 12 18 19 0 0 -74.0 48.096 -81.629 29.0 970.7
1 1 19 12 18 18 0 0 -75.0 48.300 -81.851 49.6 974.5
1 1 19 12 18 17 0 0 -76.0 48.501 -82.085 80.7 971.2
1 1 19 12 18 16 0 0 -77.0 48.685 -82.344 106.7 968.8
1 1 19 12 18 15 0 0 -78.0 48.853 -82.624 125.2 968.2
1 1 19 12 18 14 0 0 -79.0 49.023 -82.926 149.1 965.0
1 1 19 12 18 13 0 0 -80.0 49.231 -83.230 189.9 958.8
1 1 19 12 18 12 0 0 -81.0 49.486 -83.545 240.5 953.2
1 1 19 12 18 11 0 0 -82.0 49.745 -83.832 290.7 946.8
1 1 19 12 18 10 0 0 -83.0 49.971 -84.129 329.2 952.8
1 1 19 12 18 9 0 0 -84.0 50.159 -84.455 357.7 955.8
1 1 19 12 18 8 0 0 -85.0 50.351 -84.773 400.1 949.0
1 1 19 12 18 7 0 0 -86.0 50.590 -85.068 473.9 937.7
1 1 19 12 18 6 0 0 -87.0 50.882 -85.339 545.4 920.5
1 1 19 12 18 5 0 0 -88.0 51.195 -85.598 590.0 915.2
1 1 19 12 18 4 0 0 -89.0 51.505 -85.876 620.2 913.2
1 1 19 12 18 3 0 0 -90.0 51.825 -86.159 653.0 909.1
1 1 19 12 18 2 0 0 -91.0 52.147 -86.442 677.2 905.5
1 1 19 12 18 1 0 0 -92.0 52.461 -86.716 692.9 899.5
1 1 19 12 18 0 0 0 -93.0 52.758 -86.995 696.6 898.8
1 1 19 12 17 23 0 0 -94.0 53.047 -87.294 685.5 908.4
1 1 19 12 17 22 0 0 -95.0 53.320 -87.588 668.0 914.0
1 1 19 12 17 21 0 0 -96.0 53.574 -87.843 662.6 915.9
1 1 19 12 17 20 0 0 -97.0 53.832 -88.043 667.8 914.5
1 1 19 12 17 19 0 0 -98.0 54.108 -88.209 674.8 913.0
1 1 19 12 17 18 0 0 -99.0 54.396 -88.358 671.9 906.7
1 1 19 12 17 17 0 0 -100.0 54.685 -88.479 649.0 909.1
1 1 19 12 17 16 0 0 -101.0 54.950 -88.576 609.4 913.7
1 1 19 12 17 15 0 0 -102.0 55.192 -88.645 556.2 922.8
1 1 19 12 17 14 0 0 -103.0 55.420 -88.708 512.5 933.7
1 1 19 12 17 13 0 0 -104.0 55.629 -88.811 501.4 934.8
1 1 19 12 17 12 0 0 -105.0 55.823 -88.953 519.7 931.0
1 1 19 12 17 11 0 0 -106.0 56.012 -89.088 540.2 928.3
1 1 19 12 17 10 0 0 -107.0 56.193 -89.180 548.0 930.6
1 1 19 12 17 9 0 0 -108.0 56.361 -89.225 551.5 932.1
1 1 19 12 17 8 0 0 -109.0 56.515 -89.255 560.0 932.7
1 1 19 12 17 7 0 0 -110.0 56.647 -89.303 579.8 930.4
1 1 19 12 17 6 0 0 -111.0 56.750 -89.373 611.1 927.0
1 1 19 12 17 5 0 0 -112.0 56.835 -89.467 636.6 922.4
1 1 19 12 17 4 0 0 -113.0 56.921 -89.579 640.8 922.8
1 1 19 12 17 3 0 0 -114.0 57.017 -89.699 625.2 926.1
1 1 19 12 17 2 0 0 -115.0 57.112 -89.841 595.9 930.7
1 1 19 12 17 1 0 0 -116.0 57.183 -90.036 557.6 935.4
1 1 19 12 17 0 0 0 -117.0 57.228 -90.290 509.1 941.2
1 1 19 12 16 23 0 0 -118.0 57.260 -90.545 461.0 948.4
1 1 19 12 16 22 0 0 -119.0 57.285 -90.738 426.2 953.7
1 1 19 12 16 21 0 0 -120.0 57.297 -90.869 405.5 956.9
1 1 19 12 16 20 0 0 -121.0 57.304 -90.975 387.9 957.2
1 1 19 12 16 19 0 0 -122.0 57.324 -91.083 362.7 960.3
1 1 19 12 16 18 0 0 -123.0 57.354 -91.195 330.0 964.6
1 1 19 12 16 17 0 0 -124.0 57.394 -91.322 302.3 970.5
1 1 19 12 16 16 0 0 -125.0 57.444 -91.475 290.9 972.7
1 1 19 12 16 15 0 0 -126.0 57.505 -91.651 295.2 972.4
1 1 19 12 16 14 0 0 -127.0 57.574 -91.829 306.2 968.9
1 1 19 12 16 13 0 0 -128.0 57.644 -91.993 316.2 966.7
1 1 19 12 16 12 0 0 -129.0 57.715 -92.148 325.9 964.5
1 1 19 12 16 11 0 0 -130.0 57.790 -92.303 330.4 963.7
1 1 19 12 16 10 0 0 -131.0 57.871 -92.464 324.3 965.0
1 1 19 12 16 9 0 0 -132.0 57.959 -92.629 307.6 967.5
1 1 19 12 16 8 0 0 -133.0 58.051 -92.792 283.5 969.7
1 1 19 12 16 7 0 0 -134.0 58.142 -92.949 255.7 972.2
1 1 19 12 16 6 0 0 -135.0 58.231 -93.107 225.2 975.6
1 1 19 12 16 5 0 0 -136.0 58.322 -93.280 206.5 979.0
1 1 19 12 16 4 0 0 -137.0 58.428 -93.473 211.4 978.1
1 1 19 12 16 3 0 0 -138.0 58.549 -93.673 237.4 974.3
1 1 19 12 16 2 0 0 -139.0 58.670 -93.875 264.2 967.5
1 1 19 12 16 1 0 0 -140.0 58.778 -94.087 273.9 965.3
1 1 19 12 16 0 0 0 -141.0 58.875 -94.312 268.5 965.6
1 1 19 12 15 23 0 0 -142.0 58.969 -94.550 252.9 967.0
1 1 19 12 15 22 0 0 -143.0 59.057 -94.801 231.5 967.7
1 1 19 12 15 21 0 0 -144.0 59.137 -95.060 205.7 969.3
1 1 19 12 15 20 0 0 -145.0 59.209 -95.313 187.0 971.4
1 1 19 12 15 19 0 0 -146.0 59.277 -95.550 181.7 969.0
1 1 19 12 15 18 0 0 -147.0 59.337 -95.776 184.8 965.1
1 1 19 12 15 17 0 0 -148.0 59.398 -96.008 199.2 960.2
1 1 19 12 15 16 0 0 -149.0 59.481 -96.281 230.2 953.4
1 1 19 12 15 15 0 0 -150.0 59.588 -96.604 277.6 942.6
1 1 19 12 15 14 0 0 -151.0 59.683 -96.977 323.4 928.2
1 1 19 12 15 13 0 0 -152.0 59.746 -97.392 356.0 923.9
1 1 19 12 15 12 0 0 -153.0 59.777 -97.835 379.7 921.4
1 1 19 12 15 11 0 0 -154.0 59.805 -98.260 396.3 919.2
1 1 19 12 15 10 0 0 -155.0 59.846 -98.623 405.6 917.8
1 1 19 12 15 9 0 0 -156.0 59.898 -98.929 405.9 917.2
1 1 19 12 15 8 0 0 -157.0 59.963 -99.217 400.9 918.9
1 1 19 12 15 7 0 0 -158.0 60.036 -99.515 398.7 920.1
1 1 19 12 15 6 0 0 -159.0 60.113 -99.823 401.2 919.5
1 1 19 12 15 5 0 0 -160.0 60.197 -100.157 406.5 912.3
1 1 19 12 15 4 0 0 -161.0 60.291 -100.534 415.2 905.4
1 1 19 12 15 3 0 0 -162.0 60.390 -100.938 425.9 901.7
1 1 19 12 15 2 0 0 -163.0 60.488 -101.378 425.0 903.7
1 1 19 12 15 1 0 0 -164.0 60.574 -101.860 401.6 909.9
1 1 19 12 15 0 0 0 -165.0 60.647 -102.379 366.1 915.4
1 1 19 12 14 23 0 0 -166.0 60.720 -102.921 335.9 919.2
1 1 19 12 14 22 0 0 -167.0 60.769 -103.416 311.8 921.2
1 1 19 12 14 21 0 0 -168.0 60.784 -103.865 301.4 920.0SAGA GIS
Follow these instructions to install SAGA GIS
Python
Follow these instructions to install Python3
Data
Air Mass Modelling
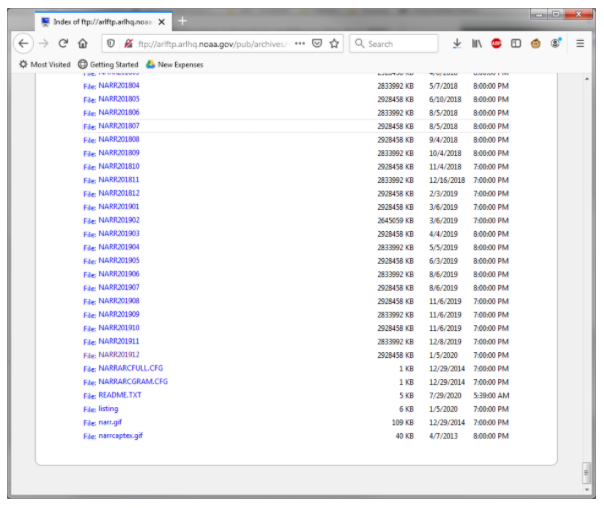
To use Hysplit, you will need information on atmospheric conditions to successfully generate a backtrajectory. Some example data are included with Hysplit. If you’d like to run your own analysis, you can download monthly data from NOAA’s ARL FTP server. This can be done with most web browsers or an FTP client. The files are named NARRYYYYMM, so for our tutorial let’s download the December 2019 file (NARR201912).
Surface Raster Data
There are a number of sources that can be used for surface water data. Here, we’ll use the General Bathymetric Chart of the Oceans (GEBCO) gridded bathymetric data. It provides depths or elevations throughout the world and is completely open to the public.
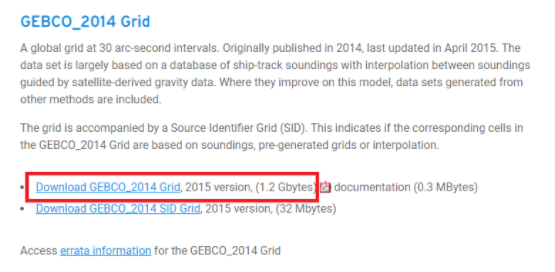
Download the GEBCO 2014 grid (or use the updated 2020 grid if you’d like but it is much larger). It will come as a NetCDF file, one of our first tasks will be to convert it to a format suitable for SAGA GIS.
Working Directory
Hysplit works best when its data files are located somewhere under the installation directory. If you won’t be using Hysplit, you can put your files anywhere. Otherwise, create a folder under your Hysplit working directory called “tutorial”. Put your NARR file in that directory.
Tutorial
Hysplit Back Trajectory Modelling (Optional)
If you’re not interested in air mass trajectory modelling, you can skip this step and use my sample “tdump” data, provided above. Save them to your working folder, then just read through the instructions so you have some sense of what the file contains.
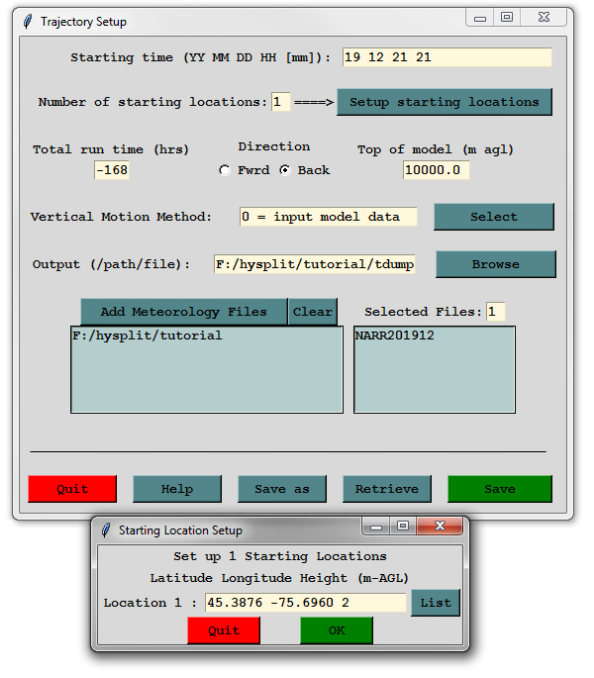
Launch Hysplit and click on “Menu”. We’re going to run a backtrajectory of the air mass over Carleton University at 2 m height on the last day of exams (Dec 21, 2019 at 9PM) for one week. Carleton University is located at 45.3876 N by 75.6960 W.
Open the Trajectory menu and select “Setup Run”. Set the parameters as follows, leave the other parameters the same
- Starting Time: 19 12 21 21
- Number of starting locations 1
- Total run time: -168
- Direction: Back
- Output path: A file called “tdump” in your working directory
- Clear the existing meteorology files and add the NARR file you downloaded earlier.
- Click the “Setup starting locations” button and enter the latitude, longitude (remember west is negative!), and starting height. Click OK, then click Save.
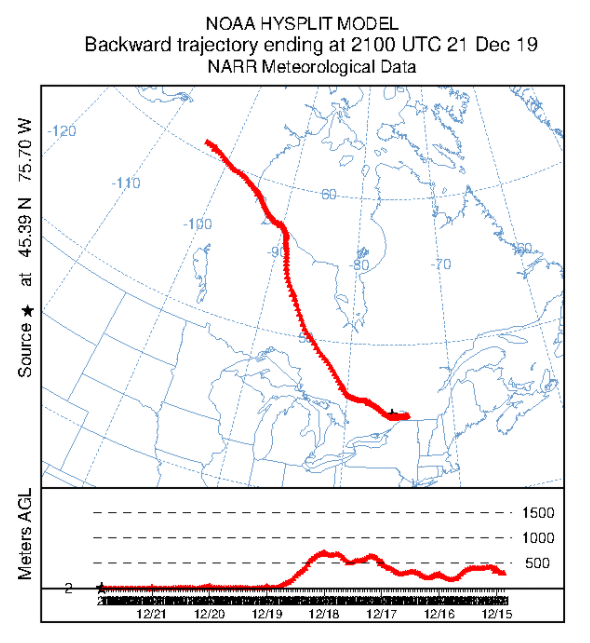
Select “Run Model” from the Trajectory model. You should see a dialog window appear and the model run. When you see “Complete Hysplit” appear, you can click the Exit button. You can see the trajectory by clicking on the Trajectory menu, the Display submenu, and then the Trajectory option. Keep all the defaults and click “Execute Display”. If you have Ghostscript / GSView set up right, you should see a map of North America and a red line indicating where the air mass travelled in the week before reaching Carleton University. The graph below shows the height of the air mass over land as it travelled.
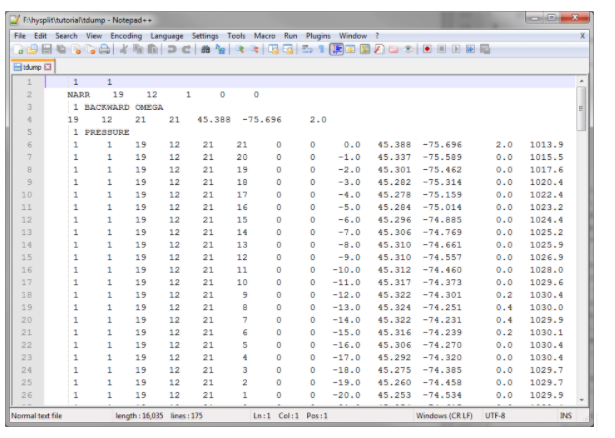
At this point, you should have a “tdump” file in your working directory. Open it up and let’s look at the contents. The first few lines tell us a bit about how the run was configured - you can see the start time, the coordinates, the input files, etc. Starting on the sixth line is the output data in space-delimited format. The columns are as follows:
- Trajectory number
- Grid number
- Year
- Month
- Day
- Hour
- Minute
- Forecast hours
- Time since start time (in hours)
- Latitude
- Longitude
- Height above ground
- Pressure
We’ll work more with these data in a few sections. Right now, let’s get our GEBCO grid ready for use.
Converting GEBCO 2014 NetCDF to SAGA GIS Grid Format
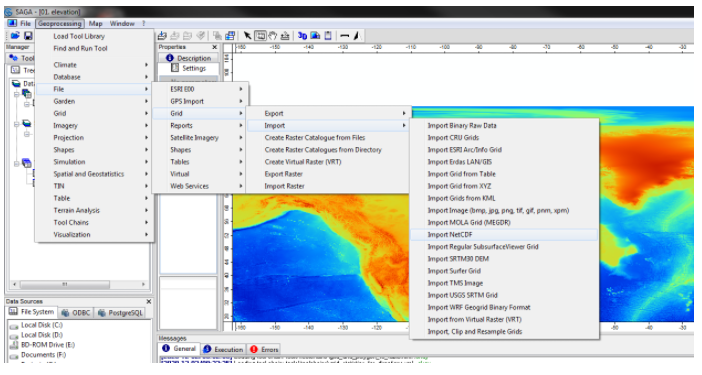
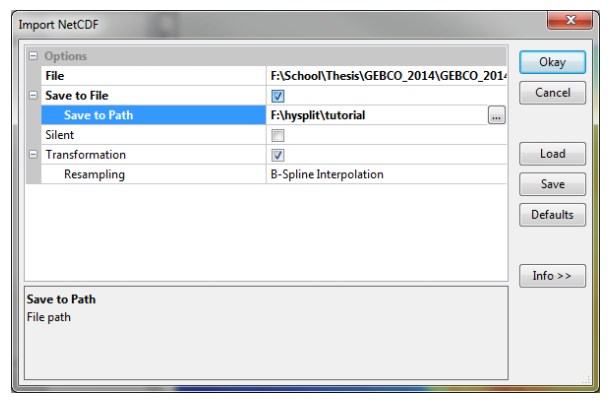
Launch SAGA GIS. While we’ll be using the command line interface for the most part, we will convert the NetCDF file using the graphical interface. Open the “Import NetCDF” tool by going to the Geoprocessing menu, then to File, Grid, Import, and finally Import NetCDF. For “File”, select the .nc file you downloaded and unzipped. Check “Save to File” and select your working directory for the Save to Path. Click Ok and let it run. When it finishes, you should have a set of files including a “.sdat” file. Make a note of the path to the .sdat file, you’ll need it later. You can then close SAGA.
Air Mass Time Over Water
To determine the time the air mass has spent over water, we will need to take several steps in both Python and with SAGA GIS. To simplify the workflow, we’ll use the Python subprocess module to call SAGA’s command line interface from within Python. The steps we will need to take are as follows:
- Extract the trajectory from the tdump file
- Segment the trajectory into sections based on the GEBCO grid
- Extract the grid value for each section. Negative values are water, positive values are land.
- Using the grid values, calculate a total time over water for the trajectory
Create a new Python file called watertime.py in your working directory.
Extracting the trajectory
To extract the trajectory, we will read the tdump file into a Python list of points. Each point will be a dictionary object with all the information about that point on the trajectory, but notably including the x and y coordinates. The Python code for this follows. Note that we skip a few lines at the beginning that correspond to data that isn’t part of the trajectory.
import subprocess
# LineSegment class is stored in lineseg.py
# from lineseg import LineSegment
# Location of your tdump file
tdump_file = r"F:\hysplit\tutorial\tdump"
# Store the points in this object
points = []
# Open the file for reading
print("Reading trajectory information")
with open(tdump_file, "r") as handle:
# split the file into lines
lines = handle.read().split("\n")
# Track the number of files
trajectory_file_count = None
# Track the number of starting points
starting_point_count = None
# Flag to skip the output type line
skip_output_type = True
# loop through lines
for line in lines:
# Split the line based on spaces and remove empty ones
data = list(filter(lambda x: not x == "", line.split(" ")))
# Skip blank lines
if len(data) == 0:
continue
# The first line we encounter is the trajectory file count
if trajectory_file_count is None:
trajectory_file_count = int(data[0])
continue
# Then we skip lines equal to the trajectory file count
if trajectory_file_count > 0:
trajectory_file_count -= 1
continue
# Repeat for trajectory points
if starting_point_count is None:
starting_point_count = int(data[0])
continue
if starting_point_count > 0:
starting_point_count -= 1
continue
# Skip the output flag line if not already done
if skip_output_type:
skip_output_type = False
continue
# Now we can read points from the trajectory
points.append({
"start_point": int(data[0]),
"grid": int(data[1]),
"year": int(data[2]),
"month": int(data[3]),
"day": int(data[4]),
"hour": int(data[5]),
"minute": int(data[6]),
"forecast": int(data[7]),
"timestep": float(data[8]),
"y": float(data[9]),
"x": float(data[10]),
"z": float(data[11]),
"p": float(data[12]),
})
print("{} points found".format(len(points)))
If your trajectory is in another format, you can adapt this method as long as the latitude, longitude, and timestep are present.
Segment the trajectory
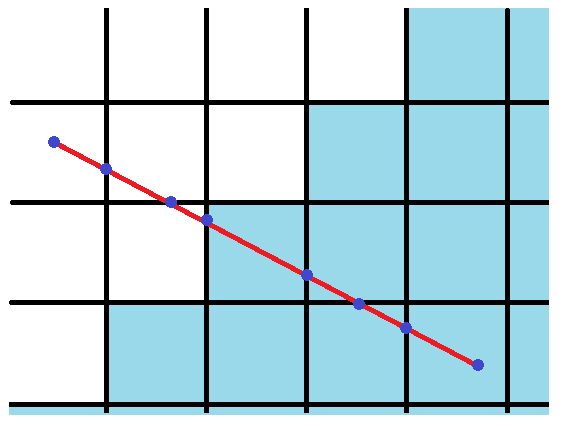
Given a grid aligned to the x- and y-axis, we can segment a straight line to pieces that extend over a single grid cell by splitting the line where it intersects either the horizontal or vertical lines of the grid. The illustration below demonstrates this by cutting the line at the blue dots.
To do this in Python is fairly straight-forward. Here, I’ve provided a class that accomplishes the task. It takes two points for the start and end of a segment and then splits it into smaller segments based on a grid with a specific origin and spacing between the horizontal and vertical lines in the grid. Save this class in a file called lineseg.py and then uncomment the import statement in the previous codeblock for the LineSegment class.
import math
class LineSegment:
# This class holds a line segment from one point to another
__slots__ = ["x1", "y1", "x2", "y2", "m"]
def __init__(self, a, b):
self.x1 = a[0]
self.x2 = b[0]
self.y1 = a[1]
self.y2 = b[1]
# slope calculation
if self.x2 == self.x1:
self.m = None # vertical line
else:
self.m = (self.y2 - self.y1) / (self.x2 - self.x1)
def gridMidpoint(self, grid_interval_x, grid_interval_y):
# Calculate the midpoint of the grid cell containing the midpoint
mp = self.midpoint()
# Calculate grid cell midpoint that contains mp
x = (math.floor(mp[0] / grid_interval_x) * grid_interval_x) + (grid_interval_x / 2.0)
y = (math.floor(mp[1] / grid_interval_y) * grid_interval_y) + (grid_interval_y / 2.0)
return (x,y)
def midpoint(self):
# Calculate the midpoint of the line segment
return ((self.x1 + self.x2) / 2.0, (self.y1 + self.y2) / 2.0)
def length(self):
# Calculate the length of the line segment
return math.sqrt(((self.y2 - self.y1) ** 2) + ((self.x2 - self.x1) ** 2))
def boundaryX(self, dx):
# Calculate the x,y coordinates of a point on the line that is dx
# away from x1
return (self.x1 + dx, (self.m * dx) + self.y1)
def boundaryY(self, dy):
# Calculate the x,y coordinates of a point on the line that is dy
# away from y1
if self.m is None: return (self.x1, self.y1 + dy)
return ((dy / self.m) + self.x1, self.y1 + dy)
def boundaryPoints(self, interval_x = 1, interval_y = 1, start_at_x = 0, start_at_y = 0):
# Build a list of points where this line intersects a grid with origin at
# start_at_x, start_at_y and with grid cells that are interval_x across and
# interval_y tall
boundary_points = [(self.x1, self.y1)]
# Handle the case where the x coordinate increases across the segment
if self.x2 > self.x1:
# Find the next intersection with the vertical lines in the grid
next_position = math.ceil((self.x1 - start_at_x) / interval_x) * interval_x
# Calculate the delta
dx = next_position - self.x1
# While the position is on the line...
while (self.x1 + dx) < self.x2:
# Add it to the list of boundary points
boundary_points.append(self.boundaryX(dx))
# Move ahead to the next grid line
dx += interval_x
# Same process, but for the case where the x coordinate decreases across the segment
elif self.x2 < self.x1:
next_position = math.floor((self.x1 - start_at_x) / interval_x) * interval_x
dx = next_position - self.x1
while (self.x1 + dx) > self.x2:
boundary_points.append(self.boundaryX(dx))
dx -= interval_x
# Same process, but for y coordinates that increase and horizontal intercepts
if self.y2 > self.y1:
next_position = math.ceil((self.y1 - start_at_y) / interval_y) * interval_y
dy = next_position - self.y1
while (self.y1 + dy) < self.y2:
boundary_points.append(self.boundaryY(dy))
dy += interval_y
# Same process, but for y coordinates that decrease and horizontal intercepts
elif self.y2 < self.y1:
next_position = math.floor((self.y1 - start_at_y) / interval_y) * interval_y
dy = next_position - self.y1
while (self.y1 + dy) > self.y2:
boundary_points.append(self.boundaryY(dy))
dy -= interval_y
# Add the end point
boundary_points.append((self.x2, self.y2))
# Sort by x coordinate to get a cohesive segment
boundary_points.sort(key = lambda x: x[0])
return boundary_points
def splitLine(self, interval_x = 1, interval_y = 1, start_at_x = 0, start_at_y = 0):
# Split a line segment into a set of segments by looking at intersections with
# a grid
new_lines = []
last_p = None
for p in self.boundaryPoints(interval_x, interval_y, start_at_x, start_at_y):
if not last_p is None:
new_lines.append(LineSegment(last_p, p))
last_p = p
return new_lines
We can then loop through the segments and split each one into a set of subsegments which correspond to the grid cells. The GEBCO 2014 grid is centered on 0 N by 0 W and has half arc-minute grid sizes. We’ll also calculate the time over that particular subsegment and put in a blank classification for now. Note that we convert the measurements from degrees to arc minutes just to make the math easier later since we have a half arc minute grid.
# Define the grid size in arcminutes
grid_x_cell_size = 0.5
grid_y_cell_size = 0.5
grid_x_start = 0
grid_y_start = 0
print("Segmenting trajectory to the grid")
last_point = None
subsegments = []
# Loop through the points
for point in points:
# If there's no last point, we're at the first point so we'll ignore it
if not last_point is None:
# Create a line segment object, converting to minutes
segment = LineSegment((last_point["x"] * 60, last_point["y"] * 60), (point["x"] * 60, point["y"] * 60))
# Determine the line segment
overall_length = segment.length()
# Determine the length of time the air was over the segment
# Absolute value since we are going backwards
# Multiply by 3600 to convert to seconds from hours.
total_travel_time = abs(point["timestep"] - last_point["timestep"]) * 3600.0
# Loop through subsegments
for subsegment in segment.splitLine(grid_x_cell_size, grid_y_cell_size, grid_x_start, grid_y_start):
# Calculate the time over this subsegment
time_over_subsegment = (subsegment.length() / overall_length) * total_travel_time
# Add it to the subsegment list with a null classification
subsegments.append({
"time": time_over_subsegment,
"line": subsegment,
"classification": None
})
last_point = point
Extracting grid values using SAGA
SAGA has a tool which will add a column to a point class with the value of a raster grid underneath the point. We’ll leverage this tool and our line segments to extract the grid values for our segments.
First, we’ll build a list of points that have to be extracted. We’ll make it a unique set of grid centerpoints to reduce the number of points we need to process. We'll also need to output the list in an appropriate format
print("Extracting unique points for depth analysis in SAGA")
extract_points = set()
# Loop through segments, find the midpoint of the grid cell that
# contains the segment, and add it to the list of points to extract
for info in subsegments:
extract_points.add(info["line"].gridMidpoint(grid_x_cell_size, grid_y_cell_size))
# We can then write out the point list to a working file
print("Creating SAGA input file")
input_point_file = r"F:\hysplit\tutorial\grid_midpoints.csv"
with open(input_point_file, "w") as handle:
# Write out the header for a CSV file
handle.write("Longitude,Latitude\n")
# Loop through and write out points
for x, y in extract_points:
# Remember that we converted to arc minutes earlier!
handle.write("{},{}\n".format(x / 60.0, y / 60.0))
Next, we’ll need to call a few SAGA commands from the command line. The three steps we’ll need from SAGA are
- Convert the CSV file into a points class (with shapes_points)
- Extract the grid values onto the points (with shapes_grid)
- Convert the points class back to something readable (with io_shapes)
To do this, we will use the subprocess module which lets us call an executable file from within Python. Here’s the first step, which creates the points class from the CSV file. Note that we tell it where it can find the SAGA executable and the CSV file, as well as the name of a new point shape file to make and that the longitude is in the first column (#0) and the latitude is in the second column (#1). We then pass the command over to the subprocess module to execute, store the output in variables, and wait for it to finish. If there was an error, we’ll exit our script at this point and you can debug your command. You should replace the file paths to the appropriate ones for your installation of SAGA and to your working folder.
# Where can we find saga_cmd.exe
SAGA_EXECUTABLE = r"C:\Program Files (x86)\SAGA-GIS\saga_cmd.exe"
print("Creating point file from table input")
# File to store output shapes in
points_shape_file = r"F:\hysplit\tutorial\grid_points.shp"
# Format the command we want with the parameters
command = "\"{}\" shapes_points 0 -POINTS \"{}\" -TABLE \"{}\" -X 0 -Y 1".format(
SAGA_EXECUTABLE,
points_shape_file,
input_point_file
)
# create a new process
p = subprocess.Popen(command, stdout = subprocess.PIPE, shell=True)
# set up variables for the output streams
(output, err) = p.communicate()
# wait for the command to complete and check the exit status
p_status = p.wait()
# if the error status isn't 0 or blank (success), we'll exit here
if not (p_status == 0 or p_status is None):
print(command)
print(output)
print(err)
print("An error occurred while running this command")
exit(1)
We’ll run the next two processes in a similar fashion, to extract the grid values and then to output it to a tab-seperated value file. Update the file paths as needed to point to your working folder and to your sdat file.
print("Extracting grid values from raster to points")
# File to store the processed shapes in
points_result_file = r"F:\hysplit\tutorial\grid_output.shp"
# Elevation file
elevation_file = r"F:\hysplit\tutorial\elevation.sdat"
# Command to extract values from grid to shapes
command = "\"{}\" shapes_grid 0 -SHAPES \"{}\" -GRIDS \"{}\" -RESULT \"{}\"".format(
SAGA_EXECUTABLE,
points_shape_file,
elevation_file,
points_result_file
)
p = subprocess.Popen(command, stdout = subprocess.PIPE, shell=True)
p_status = p.wait()
(output, err) = p.communicate()
if not (p_status == 0 or p_status is None):
print(command)
print(output)
print(err)
print("An error occurred while running this command")
exit(1)
print("Exporting shapes to tab-separated values file (TSV)")
# Location to save TSV data from the shape file to
output_file = r"F:\hysplit\tutorial\output_data.tsv"
# Command to export shapes to tab-seperated values file
command = "\"{}\" io_shapes 2 -POINTS \"{}\" -FILENAME \"{}\"".format(
SAGA_EXECUTABLE,
points_result_file,
output_file)
p = subprocess.Popen(command, stdout = subprocess.PIPE, shell=True)
(output, err) = p.communicate()
p_status = p.wait()
if not (p_status == 0 or p_status is None):
print(command)
print(output)
print(err)
print("An error occurred while running this command")
exit(1)
Next, we’ll take the tab-separated value file and convert it to a lookup table. We can use this table with the segments to find out if the values are over water (negative) or not (positive).
print("Converting TSV file to a lookup table")
grid_values = []
# Read in the TSV file
with open(output_file, "r") as handle:
# Split file contents into lines
lines = handle.read().split("\n")
# Note that we skip the header row
for line in lines[1:]:
# Split the lines based on tabs
data = list(filter(lambda x: not x == "", line.split("\t")))
# There should be at least three values at the end: long, lat, and the value from the raster
if len(data) >= 3:
grid_values.append({
"x": float(data[-3]) * 60.0, # convert back to arcminutes
"y": float(data[-2]) * 60.0,
"raster": float(data[-1])
})
Calculating Time Over Water
We’re almost done at this point. Next, we’ll need to classify the segments as land (0) or water (1) and calculate the total time over water for the whole line. Note that we could combine these steps, but they’re broken down into two steps here for clarity and in case you want to do a more complicated analysis that might require all of the segments to be classified first.
print("Classifying line segments")
# Loop through segments again
for info in subsegments:
# Find the grid midpoint
mp = info["line"].gridMidpoint(grid_x_cell_size, grid_y_cell_size)
raster_value = None
# Find the appropriate lookup point (account for floating point math errors!)
for grid_point in grid_values:
# Note that we sometimes have floating point errors due to rounding or such.
# So we'll check if the difference between the x and y coordinates of the
# lookup value is closer to the midpoint values than half the cell size.
# This should ensure that we always get the closest midpoint value
if abs(grid_point["x"] - mp[0]) < (grid_x_cell_size / 2.0) and abs(grid_point["y"] - mp[1]) < (grid_y_cell_size / 2.0):
raster_value = grid_point["raster"]
break
# Classify the segment as over water (1) or not over water (0)
info["classification"] = 1 if raster_value <= 0 else 0
print("Calculate time over water")
total_time = 0
time_over_water = 0
# Loop over the segments
for info in subsegments:
# Add time to time_over_water if it was so classified
if info["classification"] == 1:
time_over_water += info["time"]
# Keep track of the total time so we can calculate a percentage
total_time += info["time"]
# Show the user the output
# You could also do something else with it at this point, like store the
# data in a CSV file for further processing.
print("{} of {} seconds were spent over water. This represents {}% of the total time".format(
round(time_over_water, 0),
round(total_time,0),
round((time_over_water / total_time) * 100,2)
))
Running the Script
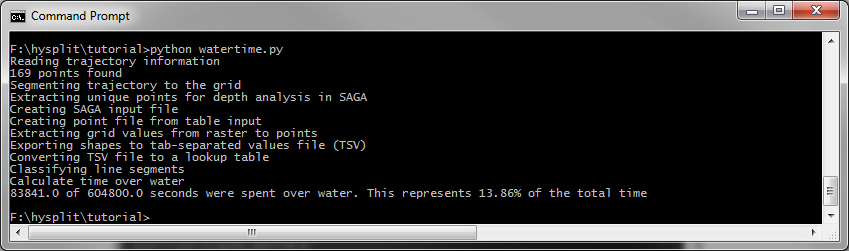
When you run it, if you have all the values set correctly, you should get output that looks like the following.
Scaling The Process
This tutorial has outlined how to accomplish this task for a single trajectory file. If you’re interested in scaling up to run many points, there are a few things you can alter for efficiency:
- First, build a list of unique grid midpoints using all of your input files and run this through SAGA. The calls to SAGA are one of the slowest parts of this process and you can save time by doing this. Keep this in a TSV file.
- Next, you can then go input file by input file, load and segment the trajectory, and perform the lookup. You can then save the output to a CSV file.
- Finally, if desired, Hysplit has a command line interface as well. You can perform a series of trajectory calculations using it and tie in the results to the above code to get a fully automated workflow for trajectory and time over water calculations.